Biological Physics
Overview
Biological Physics is an exciting interdisciplinary frontier in physics. It applies tools and techniques of physics to understand how biological systems work from the scales of single molecules to an entire living organism. At the same time, biological phenomena offer us physicists unique opportunities to learn new physics on complex, non-equilibrium systems.
The biological physics group at Carnegie Mellon combines theory, experiments and computational modeling to investigate the fundamental principles that govern the structure, mechanics and dynamic behavior of living systems across different levels of biological organization and complexities.
You can learn more about the activities in our group HERE!
See Dr. Fangwei Si's lab here:
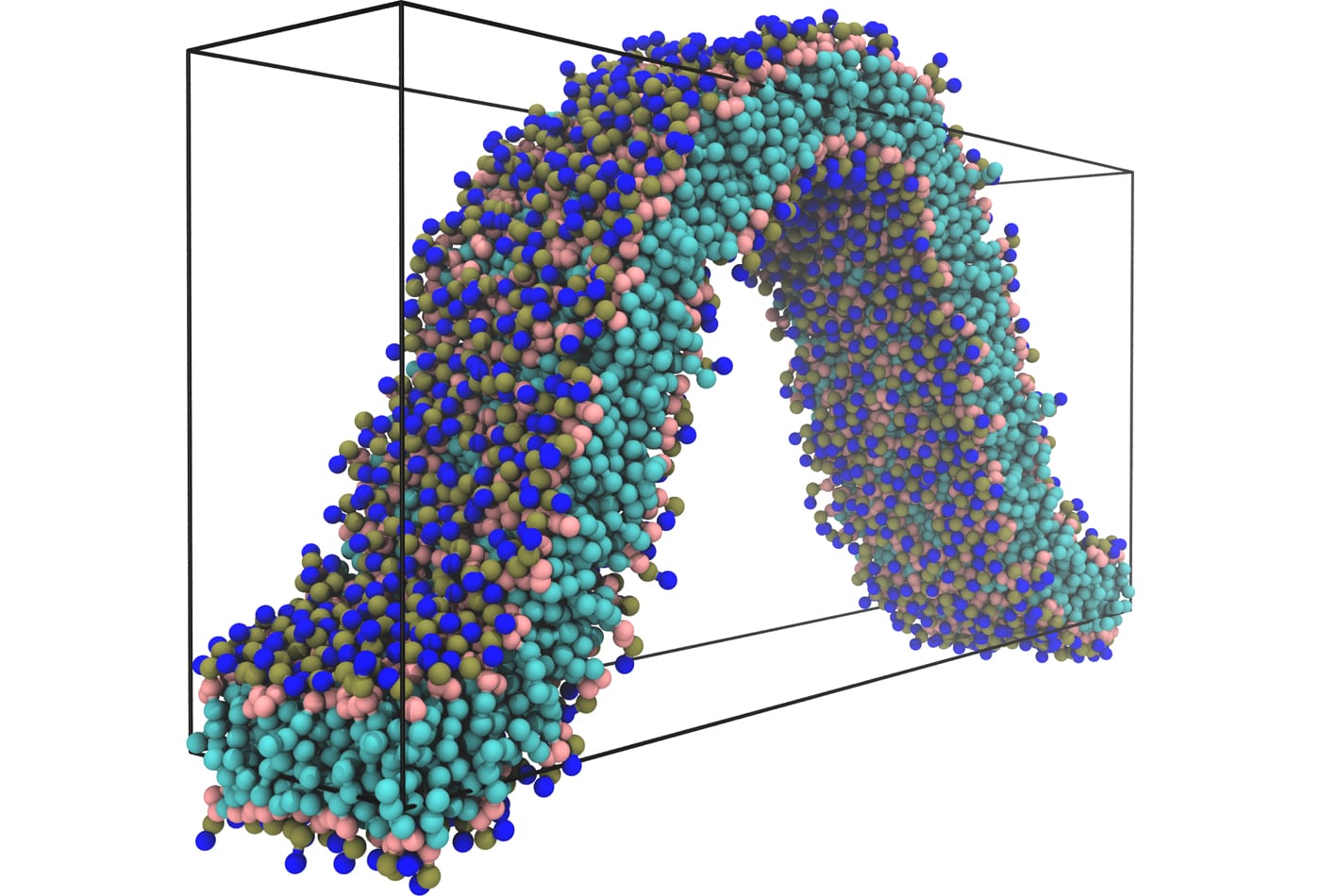
Markus Deserno uses theoretical and computational approaches – continuum elasticity theory and field thory as well as coarse-grained simulations – to study molecular-scale and mesoscale phenomena in biophysics. This allows him to study larger systems on longer time scales than in atomistic simulations and access a new arena for physical questions, many of which have biological significance. Specifically, Deserno investigates lipid membranes, proteins, viruses, or DNA on length scales larger than atomic resolution but smaller than a typical cell. On these scales, many fundamental physical concepts make a big impact on biology – among them thermal fluctuations, cooperativity, self-assembly, or elasticity. For instance, due to their surfactant-like nature individual lipid molecules in an aqueous environment spontaneously aggregate into membranes, which are laterally many orders of magnitude larger than their thickness. These quasi-two-dimensional fluid surfaces resist bending, a continuum elastic concept, but since the associated moduli are only about one order of magnitude bigger than thermal energy, membranes exhibit large thermal undulations that affect their properties.
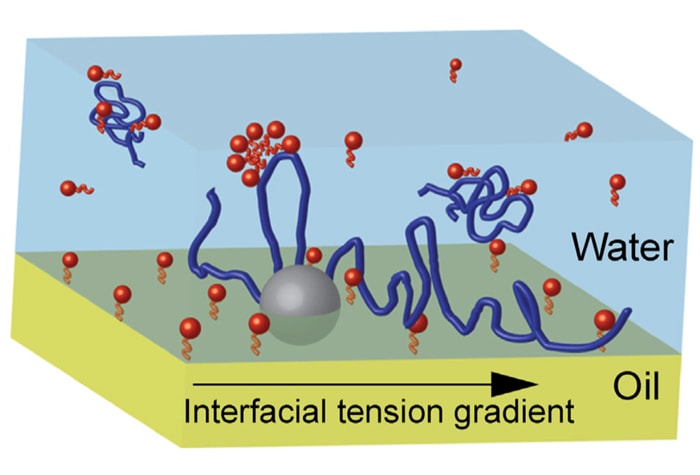
Steve Garoff and collaborators are noted for their research in complex fluids and their behavior at interfaces.Their recent research focuses on topics related to Biological and Medical Physics. In this realm, they are developing novel approaches to pulmonary drug delivery and surfactant replacement therapy by harnessing surface-induced fluid flows.
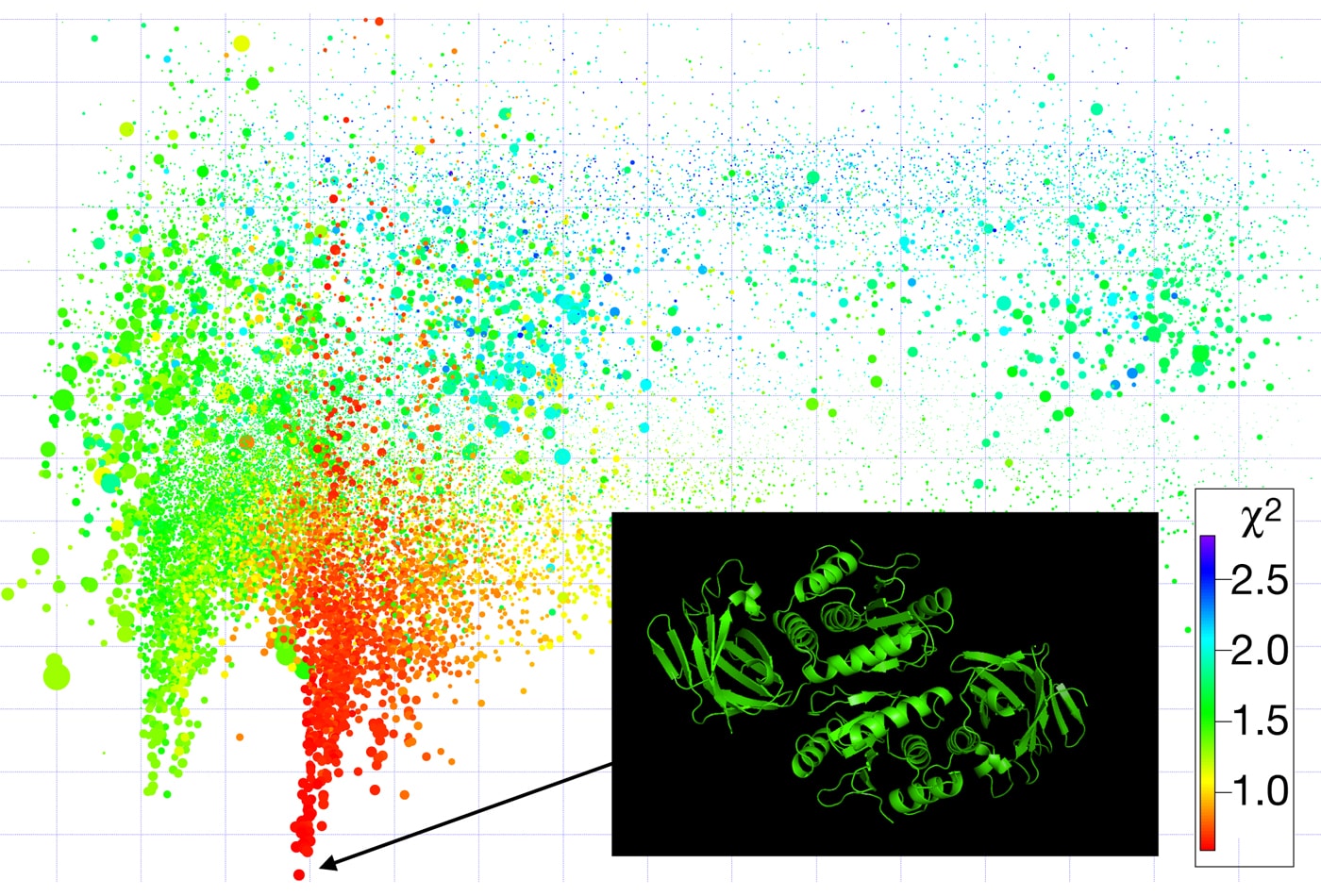
Frank Heinrich performs his research mostly at the NIST Center for Neutron Research (NCNR) and is particularly interested in disease-relevant proteins, peptides, and small molecules at interfaces and interacting with lipid membranes. Current focus is on the structural foundations of the function of the T-Cell receptor, which enables the T-Cell to recognize foreign pathogens and initiate a immune response in the body. Heinrich utilizes a broad range of surface-sensitive techniques including electrical impedance spectroscopy, surface plasmon resonance, and neutron reflectometry. At the NCNR, he is involved in the development of future-generation neutron scattering instrumentation for soft-matter and biological research and also aids academic and industrial scientists performing successful research using neutron scattering techniques.
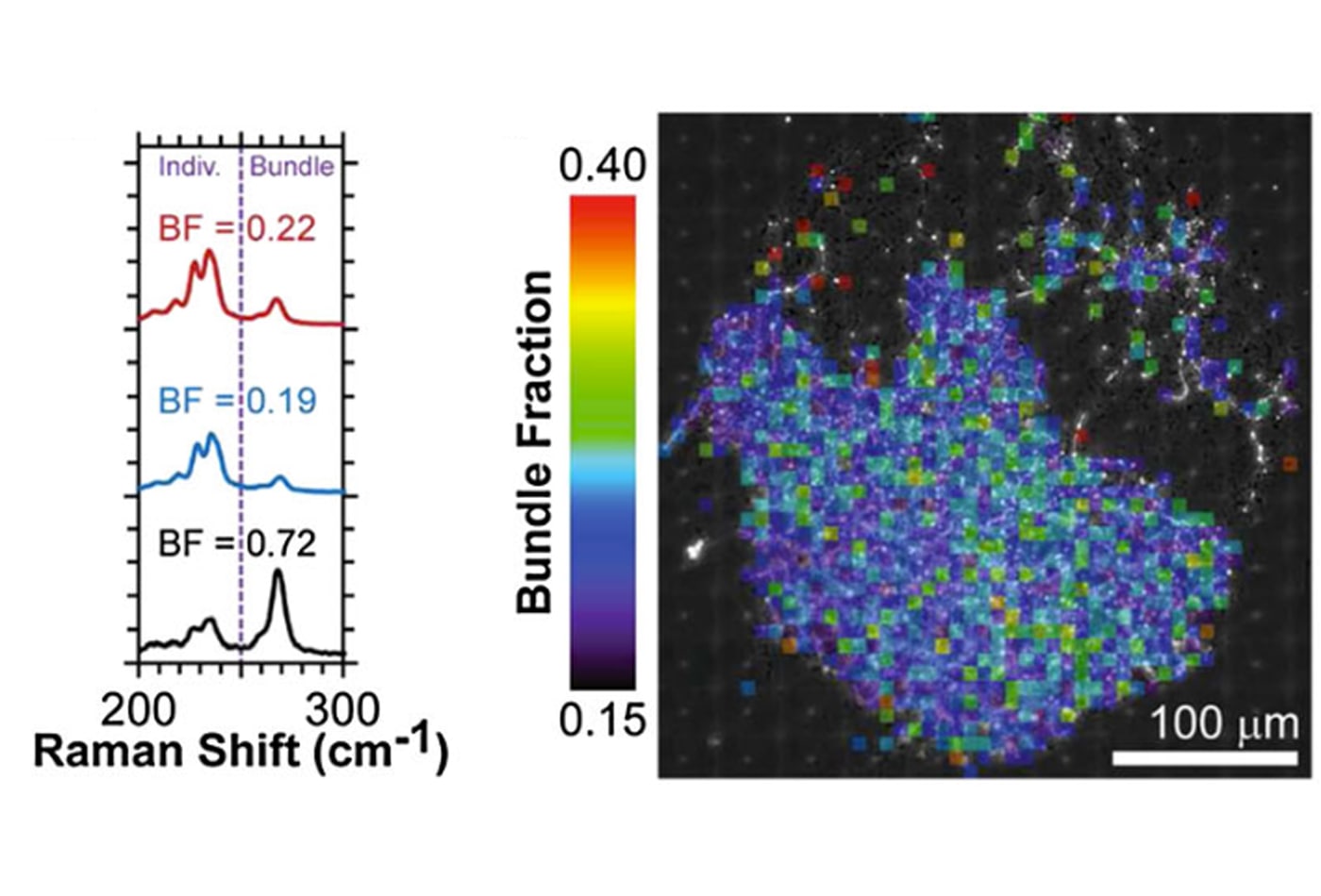
Mohammad Islam's research group employs soft matter and nanomaterials approaches to engineer multifunctional materials with tailored optical, electrical, thermal, and mechanical properties. The group also explores use of these unique materials in diverse applications such as fuel cells, supercapacitors, and drug delivery vessels and has recently focused on quantitative determination of the toxicology of carbon-based nanomaterials to biological cells.
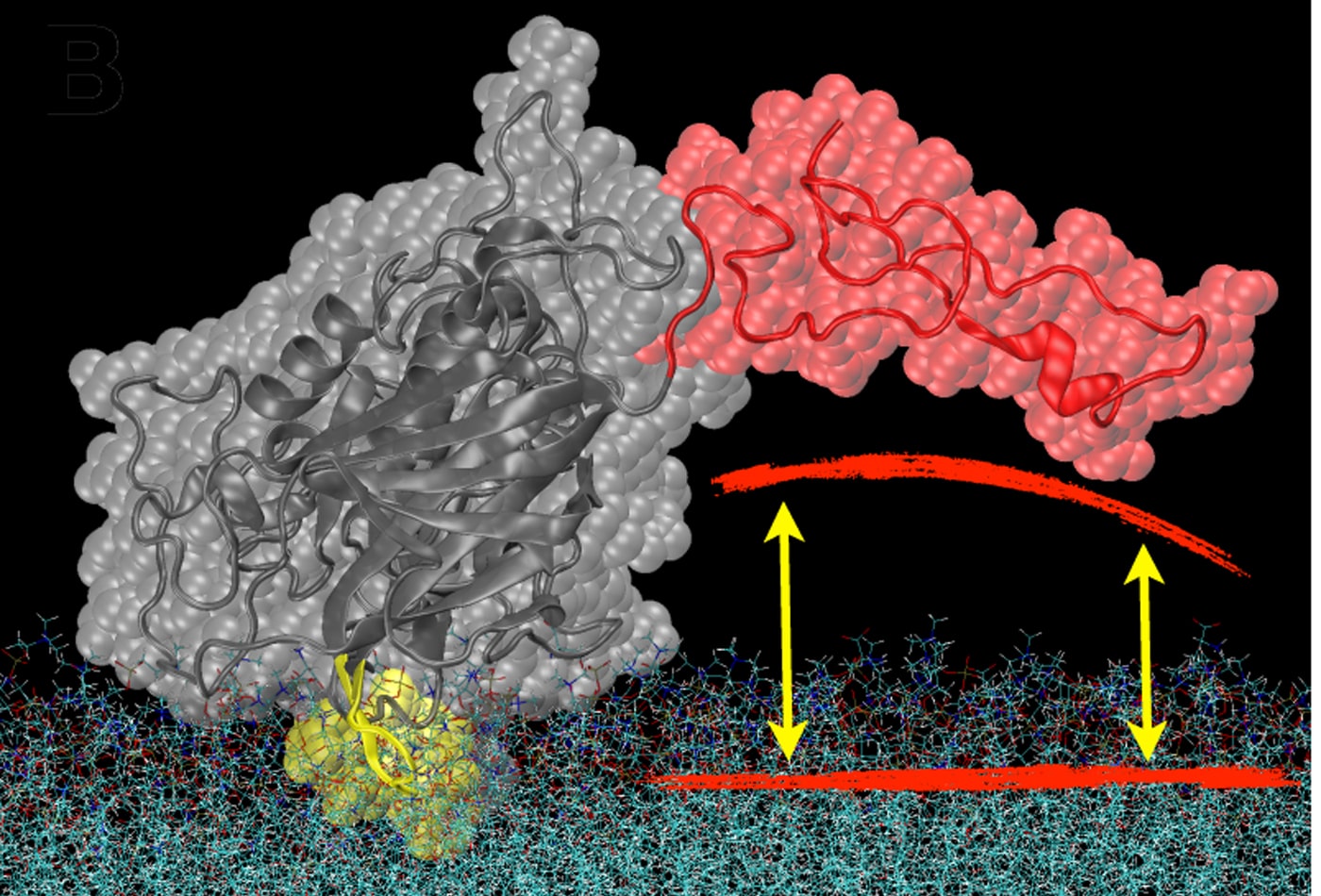
Mathias Lösche's research focuses on the functional, structural and dynamic characterization of biomimetic membranes – that is, artifical and lipid bilayer systems that are simpler than complex biological membranes but capture their fundamental Physics aspects. For example, we use surface-sensitive x-ray and neutron scattering alongside with electrochemical and advanced optical techniques to identify, characterize, and optimize membrane models in which we study the association of proteins with lipid bilayers, such as the one shown here. Microscopy techniques are used to assess the in-plane fluidity, dynamics, and 2D morphology of such systems. With this set of tools, we investigate experimentally the physical principles of membrane self-assembly and functionality. We also study the molecular origins of disease, through quantification of the membrane interactions of proteins and peptides, such as neurotransmitters, toxins or tumor suppressor proteins. These studies contribute to a deeper understanding of disease mechanisms, and help characterize cellular attack through pathogens and the self-assembly of viral particles, such as HIV, in affected cells.
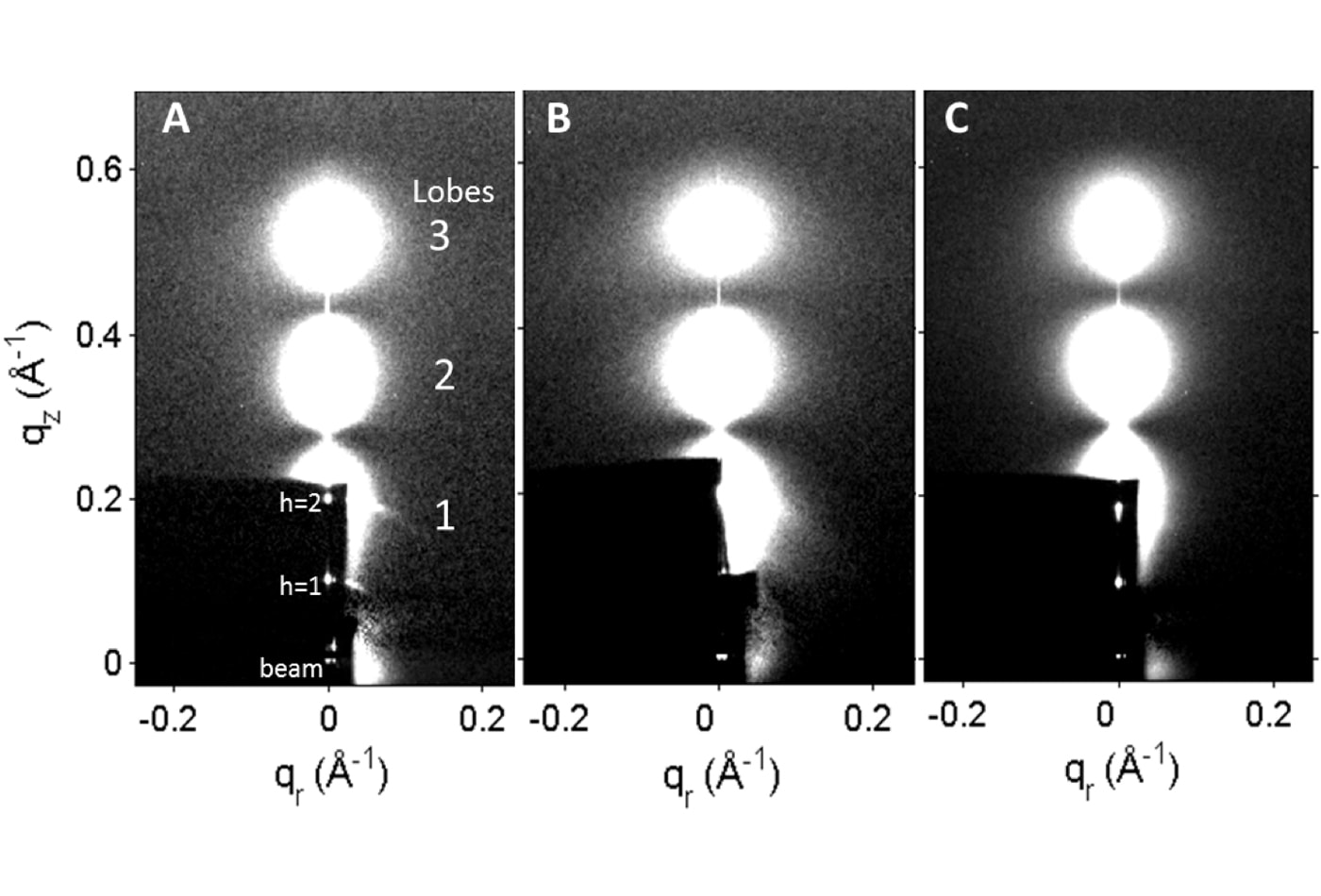
John F. Nagle is a research-active emeritus professor, well-known for his ground-breaking experimental and theoretical work on lipid bilayers, which form the structural basis of biomembranes. His studies use x-ray scattering in combination with theoretical modeling, statistical mechanical calculations and simulations, as well as volume measurements. Nagle's long-term focus has been to obtain reliable data for basic structural properties of bilayers and to elucidate the interactions between them. For biologically relevant fluid phase lipid bilayers, both issues involve measuring nanoscale fluctuations, which the group achieves using synchrotron x-rays. A longer-range goal is to understand how the molecular interactions bear out in the observed structure and the fluctuations.
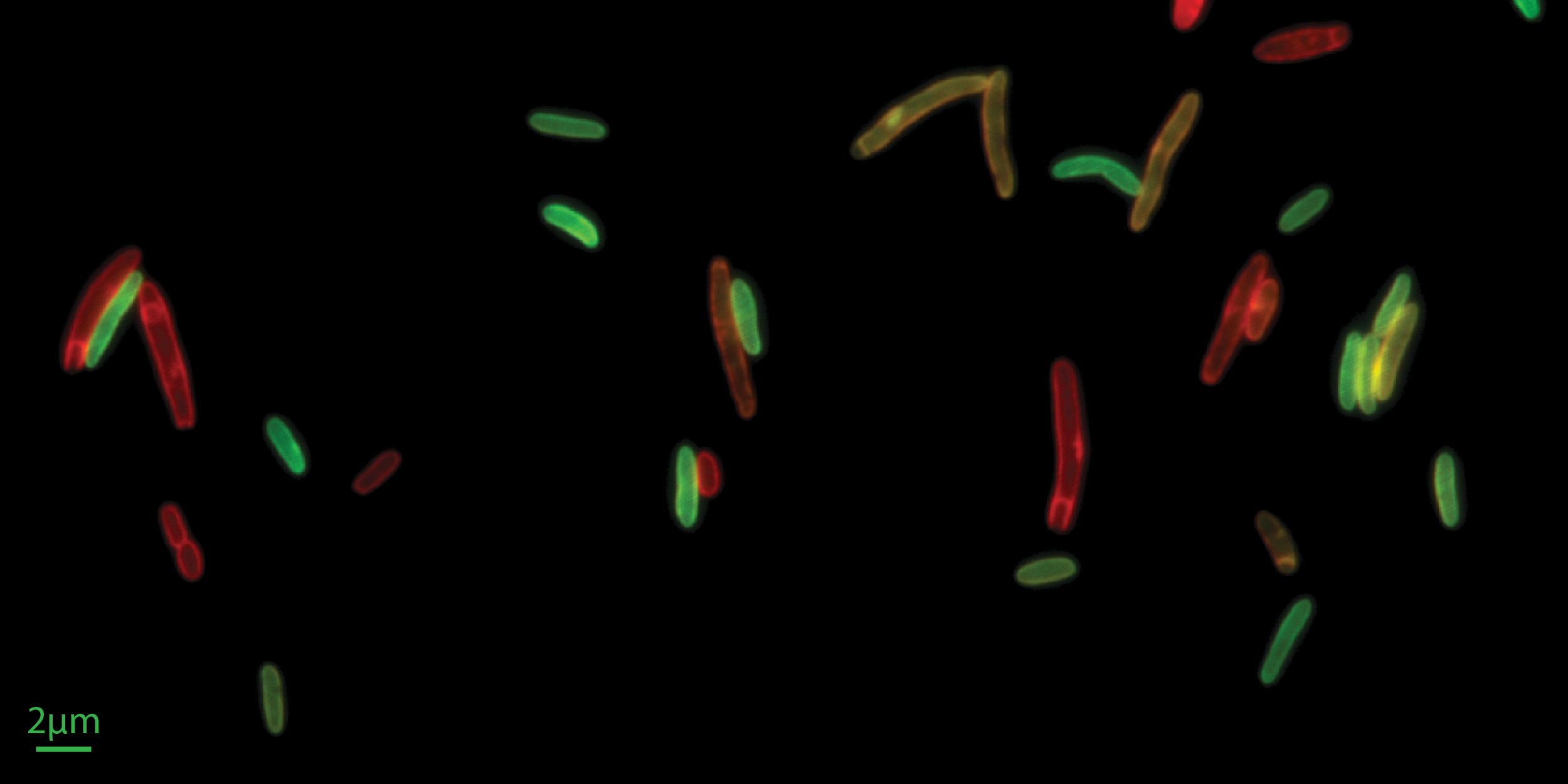
The Si lab’s drive is to discover “biological laws” that can help us understand living systems in a quantitatively precise way. Towards this goal, they develop/adapt tools, do rigorous measurements, and define new concepts. The Si lab are currently searching for simple yet fundamental rules connecting the complicated form of bacterial cells and their fitness in different environments, specifically focusing on cell surfaces and bacteria-phage interactions. The Si lab’s recent technique developments include, single-cell microfluidic tool that quantifies the binding affinity of peripheral membrane proteins in individual live cells, and membrane perturbation methods to manipulate basic properties of the membranes in individual live cells, such as membrane’s protein-to-lipid ratio and the capacity of membrane biogenesis.
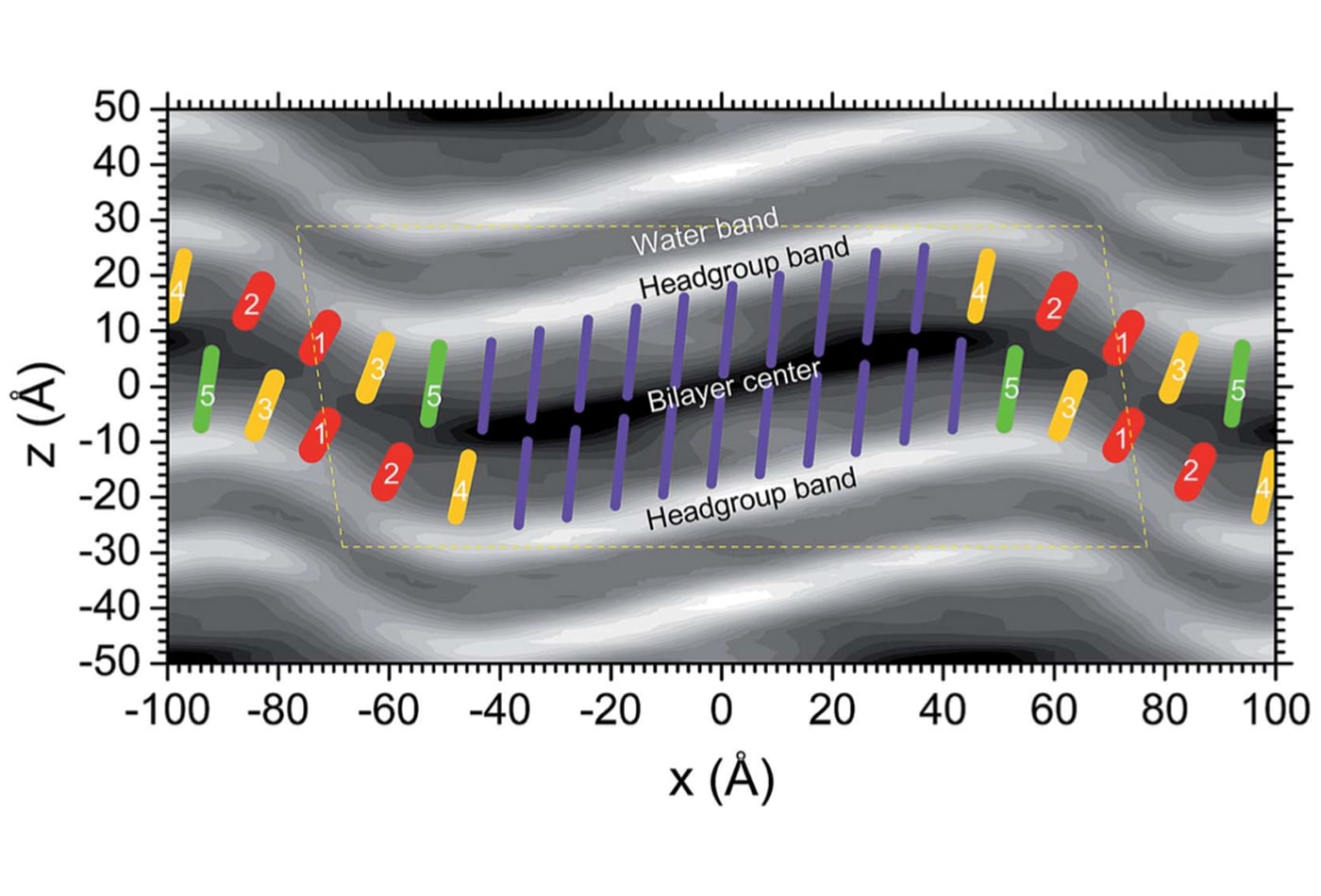
Stephanie Tristram-Nagle (emerita) performs research into lipid membrane structure, properties and thermodynamics. Her lab explores the structure of biological phospholipids, lipid/peptide and lipid/cholesterol mixtures using oriented samples on substrates and unoriented multilamellar or large unilamellar vesicles using X-ray diffuse scattering with a Rigaku rotating anode and CCD detector here at CMU, and with synchrotron radiation at the Cornell High Energy Synchrotron Source (CHESS), and her current focus is on the interaction of antimicrobial peptides with bacterial membrane mimics. The X-ray data yield not only structure, such as lipid area and membrane thickness, but also the bending and compression moduli of the membranes. Tristram-Nagle collaborates with molecular dynamics simulators to compare structural with simulated results. Another technique she uses yields lipid volume as a function of temperature by density equilibration using D2O-H2O mixtures. The group also collects volume scans vs. temperature with an Anton-Paar DMA 5000 M densitometer.