Mathias Lösche
Emeritus Professor of Physics and Biomedical Engineering (courtesy)
Associate, National Institute of Standards and Technology
Biological Physics Experiment
Supramolecular Structures Laboratory

Education & Professional Experience
Ph.D.: Technical University of Munich (Germany), Biophysics (1986)
Habilitation: Mainz University (Germany), Physical Chemistry (1994)
Professional Societies:
Fellow, American Physical Society
Member, American Physical Society (APS); Biophysical Society (U.S.); American Chemical Society (ACS); Neutron Scattering Society of America (NSSA); German Physical Society (DPG); German Biophysical Society (DGfB)
Professor of Biomedical Engineering, Carnegie Mellon University, 2008–
Professor of Physics, Carnegie Mellon University, 2005–
Director, CNBT Consortium, The NIST Center for Neutron Research, 2002–06
Research Professor (Biophysics), Johns Hopkins University, 2002–05
Associate Professor (Experimental Physics), Leipzig University, 1994–2005
Assistant Professor (Physical Chemistry), Mainz University, 1988–94
Post-doctoral Research (Biophysics): UC San Diego, 1986–88
Research Interests
In the past few decades, biology has revolutionalized our understanding of how living matter organizes itself to achieve the most astounding feat on the planet: Create intricate, organized structures that self-replicate and continuously develop further – despite the dictum of entropy which inequitably favors disorder over order. Are living systems therefore somehow miraculously exempt from the laws of physics? – Not at all! Rather, they have "learned" to exploit loopholes in the dictate of entropy in truly stunning ways! Biological physics is a fascinating branch of "mainstream physics" that tries to solve the underlying puzzles.
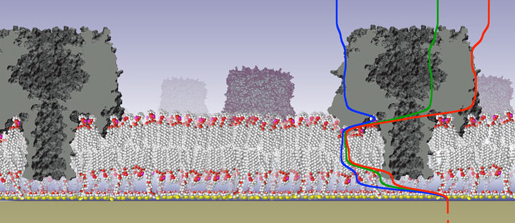
Our work in experimental biophysics focuses on the functional, structural and dynamic characterization of biomimetic membranes – that is, artifical and lipid bilayer systems that are simpler than complex biological membranes but capture their fundamental physics aspects. For example, we use surface-sensitive x-ray and neutron scattering alongside with electrochemical and advanced optical techniques to identify, characterize, and optimize membrane models in which we study the association of proteins with lipid bilayers, such as the one shown here.
Microscopy techniques are used to assess the in-plane fluidity, dynamics, and 2D morphology of such systems. With this set of tools, we investigate experimentally the physical principles of membrane self-assembly and functionality. We also study the molecular origins of disease, through quantification of the membrane interactions of proteins and peptides, such as neurotransmitters, toxins or tumor suppressor proteins. These studies contribute to a deeper understanding of disease mechanisms, and help characterize cellular attack through pathogens and the self-assembly of viral particles, such as HIV, in affected cells.
Selected Publications
B. W. Treece, F. Heinrich, A. Ramanathan, and M. Lösche, Steering molecular dynamics simulations of membrane-associated proteins with neutron reflection results, J. Chem. Theory Comput. (2020), in press. bioRxiv
M.P. Pond, R.Eells, B.W. Treece, F. Heinrich, M. Lösche, and B. Roux, Membrane anchoring of Hck kinase via the intrinsically disordered SH4-U and lengthscale associated with subcellular localization, J. Mol. Biol. (2020), in press
B. Josey, F. Heinrich, V. Silin, and M. Lösche, Association of model neurotransmitters with lipid bilayer membranes, Biophys. J. 118, 1044 (2020)
B.W. Treece, P.A. Kienzle, D.P. Hoogerheide, C.F. Majkrzak, M. Lösche, and F. Heinrich, Optimization of reflectometry experiments using information theory, J. Appl. Cryst. 52, 47 (2019)
K. Zimmermann, R. Eells, F. Heinrich, S. Rintoul, B. Josey, P. Shekhar, M. Lösche, and L. J. Stern, The cytosolic domain of T cell receptor ζ associates with membranes in a dynamic equilibrium and penetrates the bilayer deeply, J. Biol. Chem. 292, 17746 (2017)
Y. Shen, et al., PlyC, a bacteriophage endolysin that is internalized by epithelial cells and retains bacteriolytic activity against intracellular streptococci, eLife 5, e13152 (2016)
F. Heinrich, S. Chakravarthy, H. Nanda, A. Papa, P.P. Pandolfi, A.H. Ross, R.K. Harishchandra, A. Gericke, and M. Lösche, The PTEN tumor suppressor forms homodimers in solution, Structure 23, 1952 (2015)
F. Heinrich, H. Nanda, H.Z. Goh, C. Bachert, M. Lösche, and A.D. Linstedt, Myristoylation restricts orientation of the GRASP domain on membranes and promotes membrane tethering, J. Biol. Chem. 289, 9683 (2014)
F. Heinrich and M. Lösche, Zooming in on disordered systems: Neutron reflection studies of proteins associated with fluid membranes, Biochim. Biophys. Acta 1838, 2341 (2014)
S.S. Shenoy, H. Nanda and M. Lösche, Membrane association of the PTEN tumor suppressor: Electrostatic interaction with phosphatidylserine-containing bilayers and regulatory role of the C-terminal tail, J. Struct. Biol. 180, 394 (2012)
More Publications:
ORCID Researcher ID NIH-NCBI Linked In