Synthetic Molecule Invades Double-Stranded DNA
Carnegie Mellon University researchers have developed a synthetic molecule that can recognize and bind to double-stranded DNA or RNA under normal physiological conditions. The molecule could provide a new platform for developing methods for the diagnosis and treatment of genetic conditions. Their findings are published in Communications Chemistry, a new Nature journal.
The work was carried out by an international team of experts, including Carnegie Mellon Professor of Chemistry Danith Ly, an expert in peptide nucleic acid design, chemistry postdoc Shivaji Thadke and chemistry graduate student Dinithi Perera, Chemistry Professor and nuclear magnetic resonance expert Roberto Gil, and Arnab Mukherjee, a computer scientist at The Indian Institute of Science Education and Research at Pune.
“Since the double-helical structure of DNA was first elucidated by Watson and Crick, scientists have been trying to design molecules that can bind to DNA and allow one to control the flow of genetic information,” said Ly. “This is the first bifacial molecule that can invade double-stranded DNA or RNA under biologically relevant conditions.”
DNA, which contains all of an organism’s genetic information, is made up of two strands of nucleotides. The nucleotides connect with each other using hydrogen bonds, forming a helical chain of Watson-Crick base pairs. While these base pairs provide a relatively simple code to our genetic information, getting into the double helix to change the code is difficult due to the strong bonds between the base-pairs.
Ly and his colleagues at Carnegie Mellon University’s Institute for Biomolecular Design and Discovery (IBD) and Center for Nucleic Acids Science and Technology (CNAST) are leaders in the design and development of gamma peptide nucleic acids (gamma PNAs). Synthetic analogs to DNA and RNA, gamma PNAs can be programmed to bind to the genetic material (DNA or RNA) that causes disease, allowing them to search for detrimental sequences and bind to them to prevent a gene from malfunctioning.
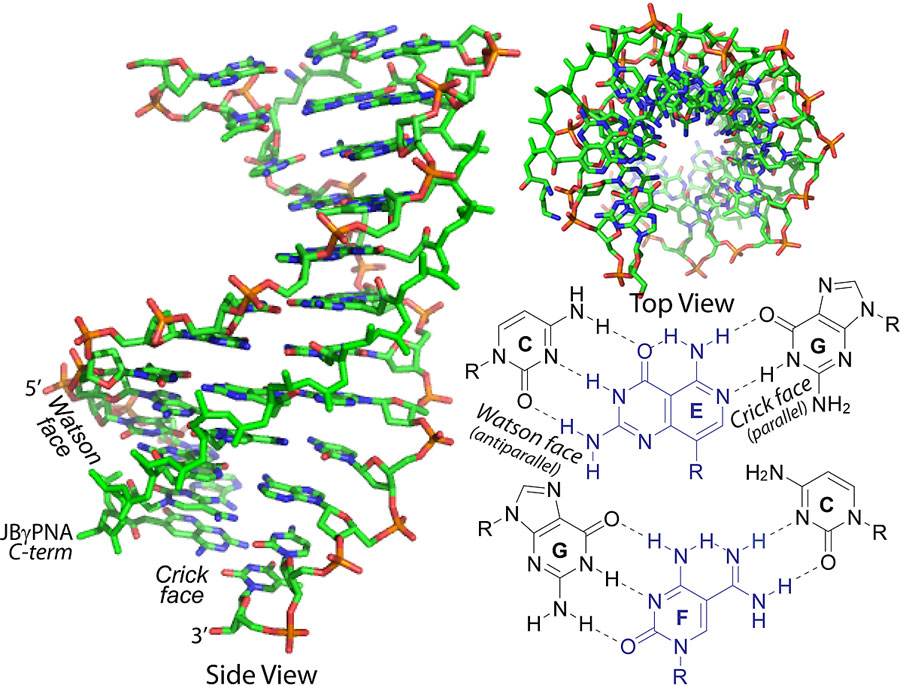
The group has created double-faced gamma PNAs called Janus gamma PNAs. Named after the two-faced Roman god, Janus PNAs are able to recognize and bind with both strands of a DNA or RNA molecule.
The concept of bifacial recognition, which is the basis of the Janus gamma PNAs, was first conceived more than two decades ago by Jean-Marie Lehn, a Nobel laureate known for his work in the field of supramolecular chemistry, and expounded on by other researchers in the field.
The advancement of this research has been held back by two obstacles. First, researchers had been able to only make a small number of Janus bases, and those bases varied considerably in shape and size. These limitations meant the different Janus bases could only recognize repeats of the same set of base pairs and couldn’t be used together like building blocks to recognize more complex sequences in DNA or RNA.
Secondly, it was difficult to synthesize Janus bases for canonical base pairs. The complementary nature of the two sides of Janus bases made the molecules hybridize and bind to each other, preventing them from incorporating into DNA and RNA.
In the current study, Ly and colleagues overcome these obstacles. They created an entirely new set of bifacial nucleic acid recognition elements, 16 in total, that accounted for every possible combination of nucleobases that could be found in the genetic code. The Janus gamma PNAs can be used to recognize any combination of base pairs and mixed and matched to detect and bind to complex genetic sequences.
Thadke solved the chemical synthesis problem by devising a novel solution- and solid-phase synthetic method to develop the Janus gamma PNAs. He also deployed a trick inherent in the helical preorganization in the backbone of the gamma PNA to prevent self-complementary Janus bases from hybridizing to one another.
These new Janus gamma PNAs have an extraordinarily high binding energy and are the first to be able to invade a canonical base-paired DNA or RNA double helix at a physiologically relevant ionic strength and temperature.
They do this by taking advantage of when double-stranded DNA and RNA molecules “breathe” and the bonds between the base pairs open for fractions of a second. When this happens, the Janus PNA inserts itself between the separated strands. If the base pairs don’t match up, the Janus PNA is ejected from the DNA molecule. But if they do match, the Janus PNA binds to both strands of the molecule.
Janus gamma PNAs have a wide-range of biological and biomedical uses. They can be designed to target genomic DNA for gene editing and transcriptional regulation. They also could be designed to bind sequence-specifically and selectively to the secondary and tertiary structures of RNA, something that traditional antisense agents and small-molecule ligands aren’t able to do. For example, the Janus gamma PNAs could be programmed to bind to RNA-repeated expansions, which could lead to new treatments for a number of neuromuscular and neurodegenerative disorders, including myotonic dystrophy type 1 and Huntington’s disease, or to noncoding RNAs, including pathogen’s ribosomal and telomerase RNA, to combat genetic and infectious diseases.
The technology is being explored by startups as well as by pharmaceutical companies for therapeutic developments.
This research was supported by grants from the National Science Foundation, National Institutes of Health and the DSF Charitable Foundation.
Originally published: https://www.cmu.edu/mcs/news-events/2018/1107_PNA-double-helical.html