Molecular Converter Switches Genetic Information from Right- to Left-Handed
Molecular Converter Could Help Advance Molecular Computers, Including Those Used to Predict Disease Outbreaks
Carnegie Mellon University researchers have developed a molecular converter that can change genetic information from right-handed to left-handed and vice versa. The converter could be an essential component for molecular computers, including one that could be used to predict infectious disease outbreaks worldwide. The findings are published in Communications Chemistry, a new Nature journal.
Molecular computing, which uses molecules like DNA as circuit components, has encountered many obstacles in its development, one being that the chemical building blocks generally can only be used in vitro. When introduced into a living host, the DNA-based components can be degraded by enzymes or inadvertently hybridize with the host’s genetic material.
In a step toward overcoming this obstacle, Danith Ly, a professor of chemistry at Carnegie Mellon, and colleagues at the Institute for Biomolecular Design and Discovery (IBD), invented a peptide nucleic acid-based molecular converter that can switch genetic information from its normal right-handed helical confirmation to a left-handed version. Researchers could use this left-handed molecule in molecular computing without having to worry about it binding with the host’s DNA.
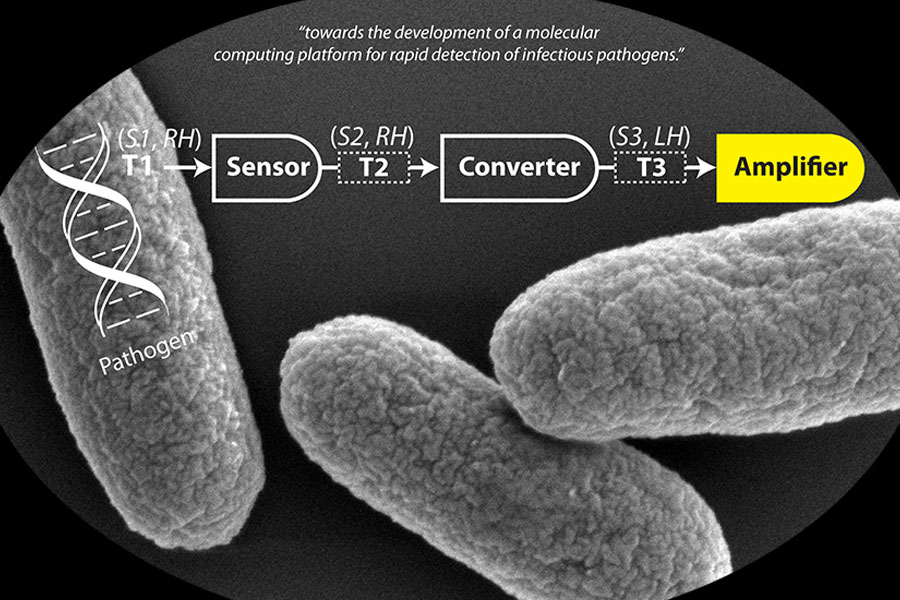
While this converter could have many applications in molecular computing, the one Ly is most excited about is its ability to be incorporated into an inexpensive and easy to use infectious disease monitoring system.
The pathogens responsible for many infectious diseases lurk, dormant throughout our environment and can be found in soil, water, insects or animals. Being able to monitor the presence of these pathogens worldwide can be critical in helping to prevent an epidemic.
However, the genetic material of pathogens can be very small, making it extremely difficult to detect pathogens in the field. Currently, to identify a pathogen, one would have to retrieve a sample and send it to a lab where the DNA or RNA could be isolated and amplified using polymerase chain reaction (PCR), prior to detection.
Ly envisions creating a tiny integrated molecular circuit (IMC) that consists of this converter, a sensor, and an amplifier. The sensor would be programmed to identify a genetic code unique to a pathogen. Someone in the field would take, for example, a water sample or a dead mosquito and add a drop of solution that contains IMC. If the selected pathogen is present, the system will detect it, convert the genetic information from a right- to a left-handed configuration and amplify the sequence. Within minutes, there would be enough material to emit a reporter signal. The person in the field could take a picture using a smart phone, and that picture could be sent to a command center, where they could determine if a pathogen is present.
“Infectious disease will continue to be a growing problem worldwide,” said Ly. “There is a misconception that infectious diseases can be eradicated. While we might be able to stop human infection, the pathogens just move from humans to another organism, or to the water or soil, where they wait for the conditions to be right for them to infect again. If we can identify when these pathogens are present in large or increasing numbers in one place, we could sound an early warning alarm and prevent an outbreak.”
Ly plans to continue working on this molecular computer, integrating the sensor, converter and amplifier. His ultimate goal is that the computer could be used one day to develop a Global Pathogen Surveillance System (GPS2). The GPS2 would monitor infectious pathogens on a global scale in real-time and play a key role in the containment and prevention of disease outbreaks.
Additional study authors include Carnegie Mellon’s Wei-Che Hsieh, Gustavo R. Martinez, Ashley Wang, Sharon F. Wu and Raunaq Chamdia.
This work was funded by the DSF Charitable Foundation, the National Science Foundation and Xunta de Galicia.
Originally published: https://www.cmu.edu/mcs/news-events/2018/1211_nucleic-acid-converter.html