Scientists Identify Achilles’ Heel of Virus’s Tough Outer Shell
 Findings Provide Potential New Target To Interfere With Viral Life Cycle
Findings Provide Potential New Target To Interfere With Viral Life Cycle
All viruses have industrial-strength shells that surround and protect the genetic material within, enabling the viral particles to remain stable, infectious and capable of spreading. Carnegie Mellon University biophysicist Alex Evilevitch and colleagues have now identified that tough shell’s Achilles’ heel.
Published in the Journal of Virology, the top journal in the field, the findings pinpoint the weakest part of the viral shell and provide a potential new target for interfering with the viral life cycle and for developing stable gene therapy delivery vehicles.
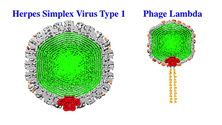
The outer shell, called a capsid, encases a virus’s genetic material. Viruses like Herpes Simplex virus type 1 (HSV-1) contain double-stranded DNA, the long strands of which are tightly packed and exert a tremendous amount of pressure, reaching tens of atmospheres, on the interior capsid wall. In previous work, Evilevitch measured for the first time this pressure in HSV-1; he also has shown that it is this pressure that propels DNA out of a small portal in the virus’s capsid and into a host cell.
With his latest research, Evilevitch and physics graduate student David Bauer reveal that the portal is more than just a conduit for DNA. It also is the weakest structural part of the capsid.
“The most exciting thing here is that we’ve shown, for the first time, how DNA pressure affects portal stability, which ultimately determines the virus’s stability over time at any temperature,” said Evilevitch, associate professor of physics and a member of CMU’s Center for the Mechanics and Engineering of Cellular Systems.
The portal is a critical component of viral capsids. Made up of several different proteins, the portal complex actively packages DNA during viral assembly, releases DNA during infection and, as the new research reveals, is key to maintaining the capsid’s delicate balance of being stable enough to retain the genome while being unstable enough to allow efficient release of the genome during infection.
“Previous experiments have investigated the role of internal pressure on the structural integrity of viral capsids,” Evilevitch said. “Here we provide the first experimental evidence that it is the mechanical strength of the portal complex itself that determines virus stability with regard to genome retention.”
For this study, Evilevitch and his team looked at portal complex stability in three different, viruses — two viruses that infect bacteria (Lambda and P22) and one that infects human cells (HSV-1). All three viruses contain double-stranded DNA that exerts pressure on the capsid wall.
Using a novel differential scanning microcalorimetry assay they developed, the researchers heated the virus samples and detected the temperature at which the portal opens to release DNA. That temperature reflects the mechanical stability of the portal.
For each type of virus they studied, the researchers created mutant strains of that virus by varying the length of packaged DNA within, thus creating mutants with different internal pressures. The results of the assays revealed that mutants with more packaged DNA and therefore more pressure, released their DNA through the portal at a lower temperature. This finding suggests that the mechanical force of the genome pushing against the virus’s portal is destabilizing it and making it prone to breaking open.
Additionally, the research team compared the temperature of DNA release for the three different virus types. The results revealed an increase in the portal stability for viruses that have longer genomes and therefore have higher pressures inside.
“Our results suggest that the portal complex has evolved to withstand the outward force of the packaged genome balanced against the requirement of efficient DNA release during infection,” Evilevitch said. “Further understanding of this balance between internal pressure and portal stability offers new insights for interfering with viral replication, as well as designing viral vectors for gene therapy that can stably retain foreign nucleic acid.”
In addition to Evilevitch and Bauer, researchers involved with the project include: CMU’s Dong Li; the University of Pittsburgh School of Medicine’s Fred Homa and Jamie Huffman; the University of Utah School of Medicine’s Kasandra Wilson, Justin Leavitt and Sherwood Casjens; and Louisiana State University School of Veterinary Medicine’s Joel Baines.
This research was funded by the Swedish Research Council and the National Science Foundation (CHE-1152770 to AE) with additional support from the Public Health Service grants and a National Institutes of Health training grant.
By: Jocelyn Duffy, jhduffy@andrew.cmu.edu