
Dr. Amir Barati Farimani
Associate Professor, Mechanical Engineering and Biomedical Engineering
- Scaife Hall 310
- 412-268-1997
- 412-268-3348
Scaife Hall 310
Carnegie Mellon University
5000 Forbes Avenue
Pittsburgh, PA 15213
Education
- B.S. Mechanical Engineering, Ferdowsi University of Mashhad, 2005
- M.S., Mechanical Engineering, Tehran TMU University, 2008
- Ph.D., Mechanical Science and Engineering, University of Illinois at Urbana-Champaign, 2015
Bio
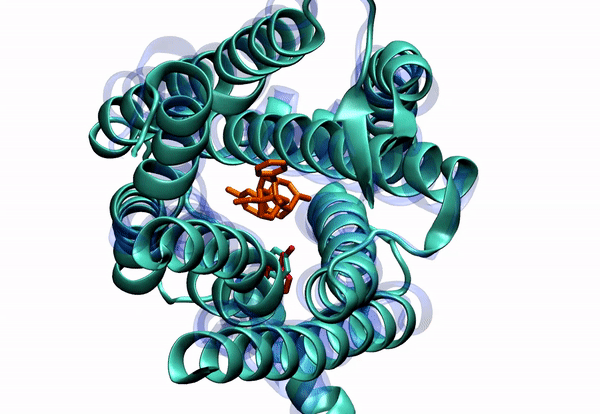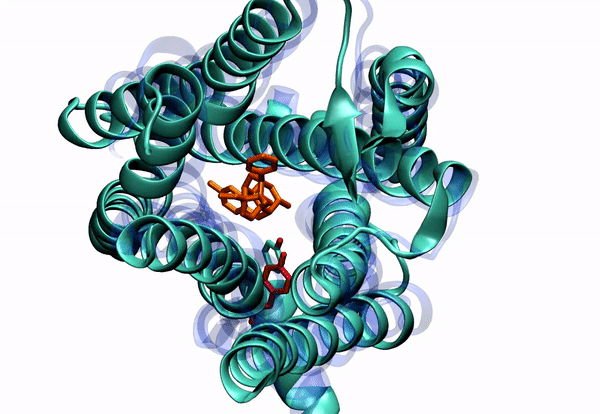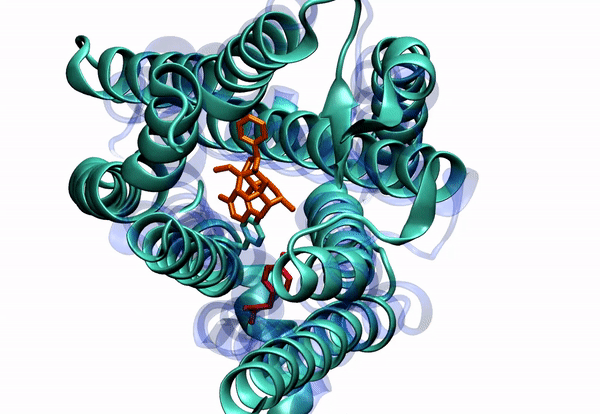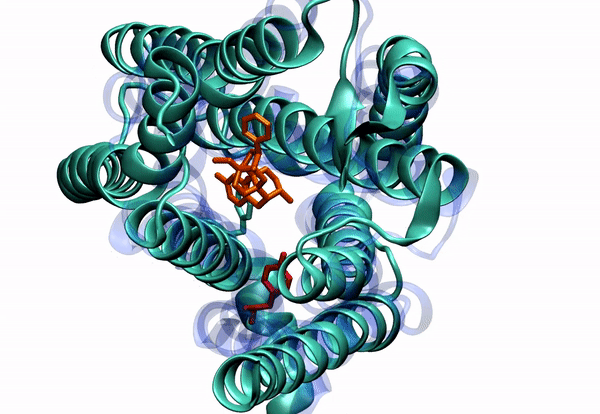
Dr. Barati Farimani received his Ph.D. in 2015 in Mechanical Science and Engineering from University of Illinois at Urbana-Champaign. His PhD thesis was about "Detecting and Sensing Biological Molecules using Nanopores". He extensively used atomistic simulations to shed light on the DNA sensing and detection physics of biological and solid state nanopores. Right after that, he joined Professor Vijay Pande lab at Stanford. During his post-doc, he combined machine learning and molecular dynamics to elucidate the conformational changes of G-Protein Coupled Receptors (GPCRs). He specifically was focused on Mu-Opioid Receptors to elucidate their free energy landscape and their activation mechanism and pathway.
The Barati Lab at Carnegie Mellon University is broadly interested in the application of machine learning, data science and molecular dynamics simulations to health and bio-engineering problems.
Research
The main theme of our lab is doing research at the interface of bioengineering, machine learning and computation. We are combining molecular dynamic simulations, machine learning and statistical learning to understand and predict the properties and interactions of bio-molecules. To be more specific, we are focused on two types of problems:
1. Interactions and recognition of bio-molecules such as DNA with synthetic materials: We perform MD simulations to generate the trajectories. We use statistical learning techniques to learn the collective variables. This is an interesting area where statistical mechanic and machine learning coincides.
2. To understand small molecule and protein interaction. We are dealing with super high dimensional time-series data coming from MD simulations. Reducing the dimension of the data, and learning the reaction coordinates help us understand many fundamental biophysical and physiological mechanisms for protein conformation and dynamics.
Research Interests: MD simulations, statistical learning
Awards and Recognition
-
Stanley I Wise Best Thesis Award, University of Illinois, 2015