New Insights into Molecular Mechanisms Ensuring Proper Assembly of Ribosomes
Large RNA-protein complexes called ribosomes decode the genetic information contained in messenger RNA and catalyze protein synthesis. These molecular machines are central to cellular metabolism, growth, and survival. In eukaryotes, about 200 accessory proteins known as assembly factors facilitate ribosome assembly. Despite their central role in cellular processes, how assembly factors help to build ribosomes is poorly understood.
In the Woolford lab, recent alumna Salini Konikkat took on research aimed at understanding the mechanism of action of an assembly factor, Erb1. Her research was published in the journal Nucleic Acids Research1, authored by Konikkat, John Woolford, and current graduate student Stephanie Biedka.
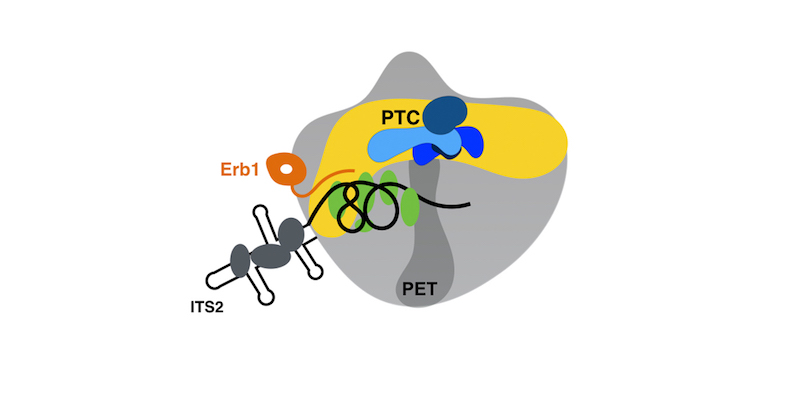
(Above image: Cartoon illustration of a 60S pre-ribosome (grey) with the peptidyl transferase center, PTC (yellow), that catalyzes protein synthesis, and the tunnel (dark grey) through which the nascent polypeptides exit. While studying Erb1, Konikkat discovered that the assembly of proteins or RNA around the PTC (blue) and PET (green/black) are required for removal of the ITS2 spacer RNA, an irreversible step required to build mature 60S subunits. This newly discovered mechanism ensures proper assembly of the active centers on ribosomes, and prevents cells from making spurious ribosomes.)
Instead of taking a traditional approach that involves the depletion of the entire protein from cells, Konikkat screened for functionally-defective Erb1 proteins capable of entering pre-ribosomes. This tactic allowed the lab to identify new roles for Erb1 in 60S ribosomal subunit assembly, and can be applied to examine functions of other assembly factors. Using proteomic and biochemical experiments, Konikkat showed that functional centers of ribosomes involved in protein synthesis, namely the peptidyl transferase center and the polypeptide exit tunnel, fail to assemble properly in these mutants. These misassembled ribosomes containing defective Erb1 fall apart, and do not undergo further maturation.
Observations from this study point to a mechanism by which functional centers on ribosomes are examined for proper assembly inside the nucleus, before they exit into the cytoplasm, the site of protein synthesis. Konnikat also discussed the implications of these observations to the ribosome assembly field in a review article describing the principles and molecular details of 60S ribosomal subunit assembly, "Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast," which was published in the Biochemical Journal2.
1. Konikkat S, Biedka S, Woolford JL Jr. The assembly factor Erb1 functions in multiple remodeling events during 60S ribosomal subunit assembly in S. cerevisiae. Nucleic Acids Res. 2017 Jan 23. pii: gkw1361.
2. Konikkat S, Woolford JL Jr. Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem J. 2017 Jan 15;474(2):195-214.

