Intercultural Communication: The Secret to Pneumococcal Success?
Bacterial cells communicate with each other via specialized signaling systems. These signaling systems are pivotal for pathogens to sense and respond to changes in host environment. Activation of signaling pathways induces changes in gene expression and phenotype, and ultimately promotes either asymptomatic colonization or disease and can underlie decreased susceptibility to clinical interventions.
In a new research study from the Hiller lab published in PLoS Pathogens1, Anagha Kadam, a fifth-year Ph.D. student and a recipient of the Glen de Vries Presidential Fellowship, and colleagues present the discovery and characterization of a new signaling system in Streptococcus pneumoniae (pneumococcus). Pneumococcus is an opportunistic pathogen that can either asymptomatically colonize the human throat or disseminate to tissues and cause severe respiratory or systemic diseases; it is one of the top human pathogens and worldwide causes over one million annual deaths in young children and the elderly.
This novel signal serves in messaging across pneumococcal cells, analogous to a new word in the lexicon of bacterial communication. The signaling system consists of a transcriptional regulator and a secreted peptide. Using extensive molecular tools and the mouse model of pneumococcal disease, Kadam and colleagues show that this system promotes commensalism over lung disease. Further, they find that this signaling system is unique to a group of pandemic, multi-drug resistant pneumococcal strains called the PMEN1 lineage. This ability to carefully balance asymptomatic colonization versus systemic disease may have contributed to the worldwide spread and success of this pandemic lineage.
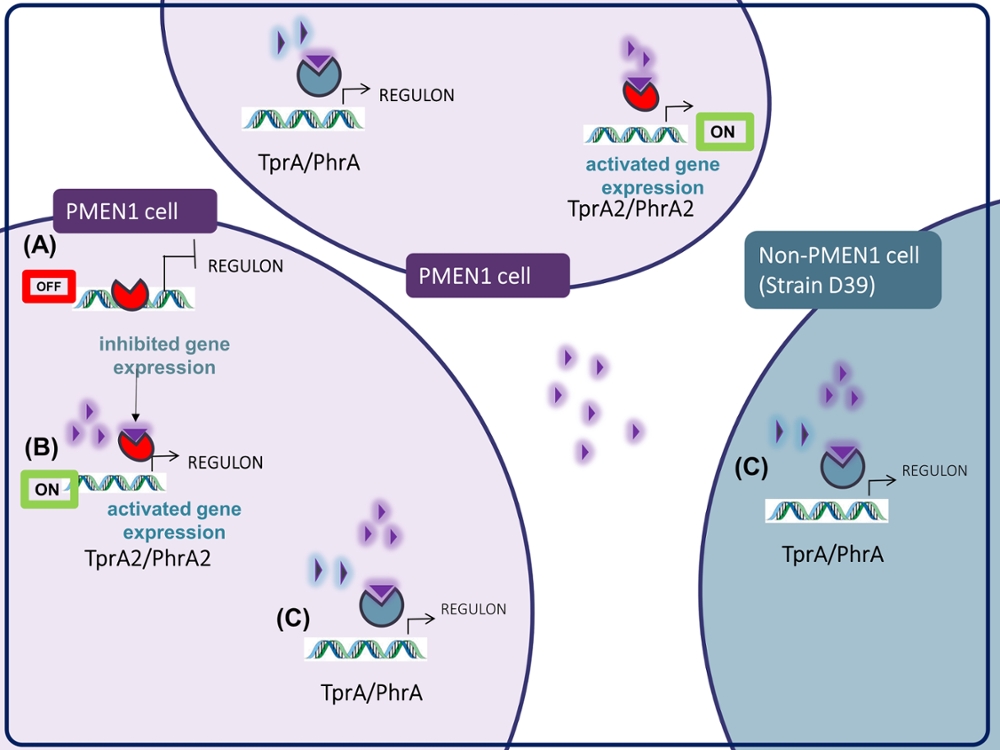
In an exciting twist, while this unique communication signal is emanated by only the pandemic PMEN1 strains, it can be received by the non-PMEN1 strains in the neighborhood and manipulate a second signaling system that plays a role in disease. This finding is important in the context of prevalent multi-strain infections, wherein the presence of PMEN1 strains may tune the virulence potential of neighboring strains and impact disease outcome in the host. That is, strains may behave differently when in the presence of PMEN1. This work reveals a new code of communication employed by a clinically relevant lineage and opens new avenues for the study of inter-strain interplay during infection.
Read more about the work on Medical Xpress.
1. Kadam A, Eutsey RA, Rosch J, Miao X, Longwell M, Xu W, et al. (2017) Promiscuous signaling by a regulatory system unique to the pandemic PMEN1 pneumococcal lineage. PLoS Pathog 13(5): e1006339. DOI: 10.1371/journal.ppat.1006339

