New Names for Old Gene Transfers
Research by Dannie Durand and her students fills a gap in a widely-used framework for classifying the evolutionary relationships between genes, reported1 in the March 1, 2017, issue of Bioinformatics.
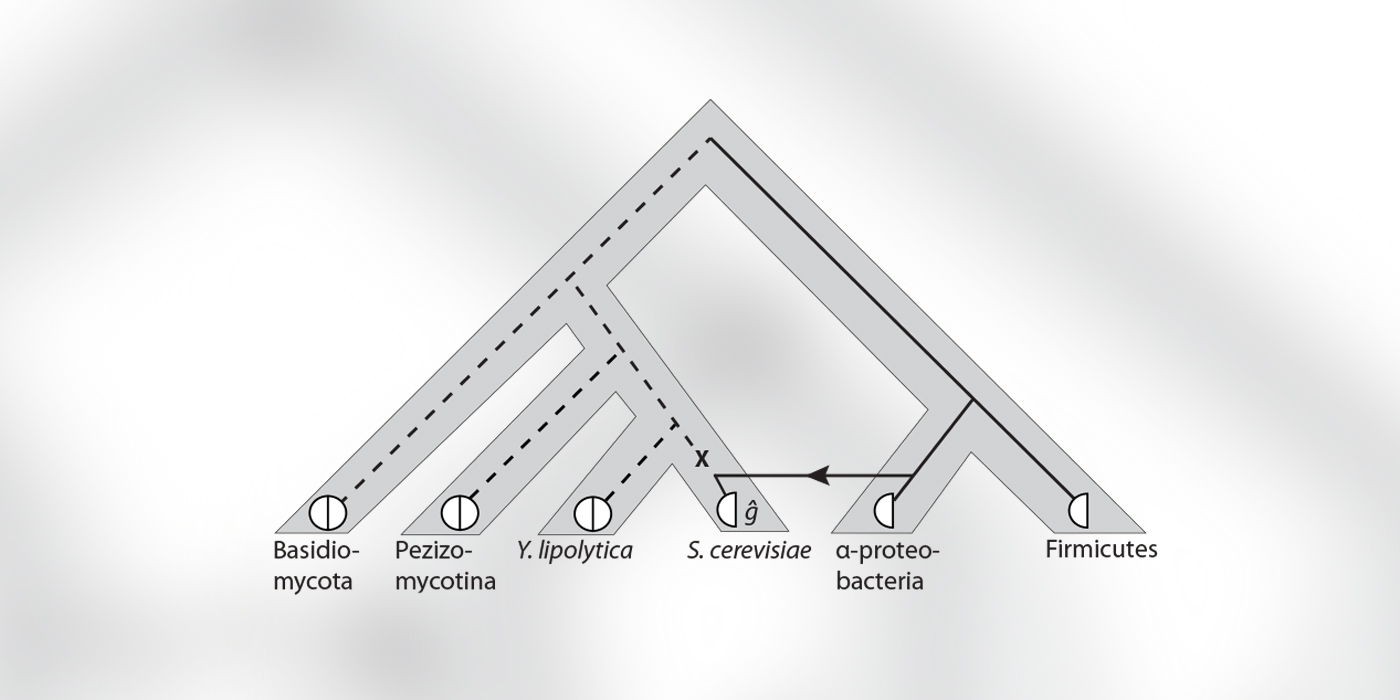
Nature makes new genes by copying and modifying existing genes. As a result, genes that share common ancestry tend to have similar functions. Understanding how genes are related provides important clues to what they do. In fact, function prediction based on evolutionary information is so powerful that a framework has been developed for classifying gene pairs based on their shared evolutionary history. This framework is fundamental for basic research and bioinformatics applications in comparative genomics. It is the basis for identifying candidate genes in model organism studies and an essential component of pipelines to annotate genes in newly sequenced genomes.
The existing framework classifies genes related through speciation and gene duplication. However, it cannot handle genes that were acquired from another organism by horizontal transfer, the primary way that bacteria obtain new genetic material that allows them to adapt to new challenges. To meet this need, Dannie Durand and her students developed a principled framework that also considers horizontally transferred genes. Their new framework offers precise, unambiguous definitions that are mutually consistent and preserve the properties of the classification framework currently in use. An algorithm realizing these definitions has been implemented in the Durand Lab's publically available Notung software package.
This is one of many examples of biological sciences students at all levels making scientific contributions through original research that is published in peer-reviewed journals. The new framework was jointly developed by former Carnegie Mellon student Charlotte Darby and postdoctoral researcher Maureen Stolzer. Charlotte obtained B.S. and M.S. degrees in computational biology from Carnegie Mellon and is now a first year doctoral student at Johns Hopkins. Maureen obtained her Ph.D. in biological sciences and a secondary master's degree in machine learning, both from Carnegie Mellon. Patrick Ropp, a graduate of Carnegie Mellon's master of science program in computational biology, contributed a practical demonstration of the approach by applying the framework to approximately 13,000 bacterial gene families.
1. Charlotte A. Darby, Maureen Stolzer, Patrick J. Ropp, Daniel Barker, Dannie Durand; Xenolog classification. Bioinformatics 2017; 33 (5): 640-649. doi: 10.1093/bioinformatics/btw686



