CRIME Pays... in Genome Science!
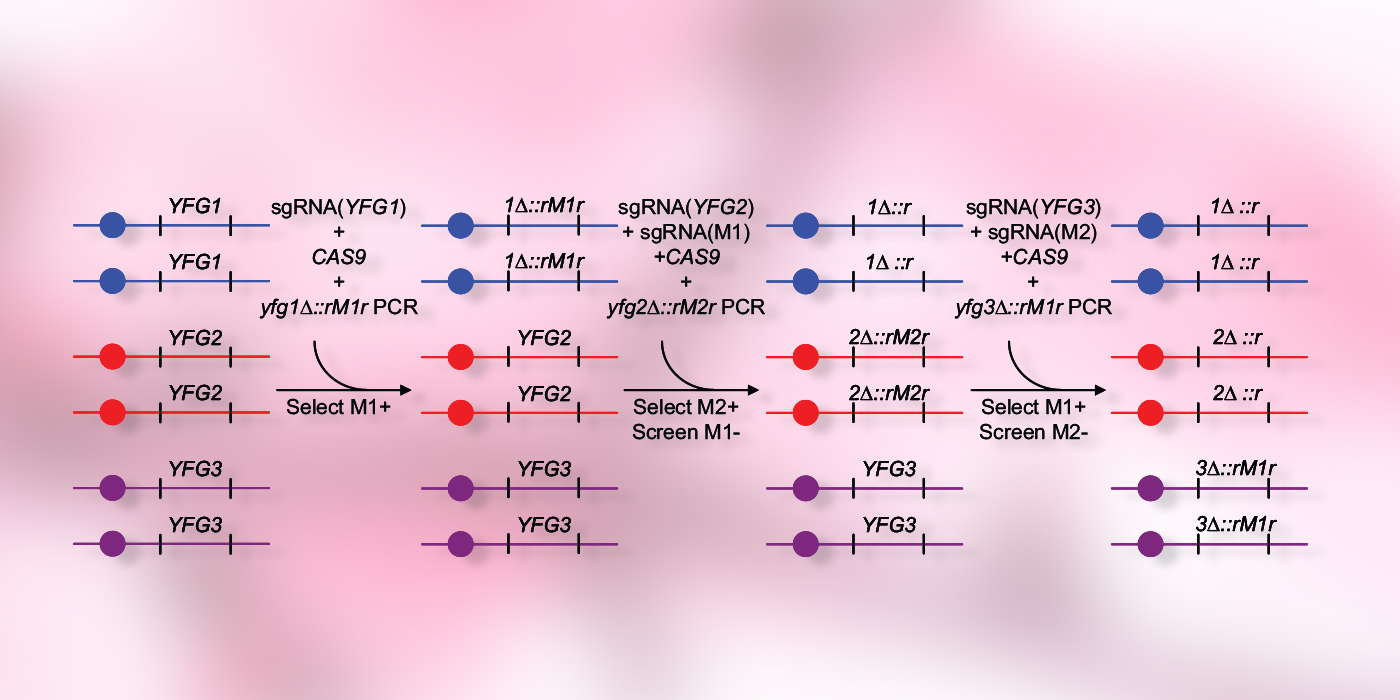
Our ability to modify genomes is limited by the number of selection markers. These markers allow genome scientists to isolate rare cells with a specific designer genome alteration from a large population. Hence a genome scientist typically uses three markers to find a cell with three alterations, and if you do not have three markers you are out of luck! Therefore, the selection marker limitation constrains genome science.
"Marker recycling" is a strategy developed by 20th century geneticists to overcome the selection marker limitation. The strategy allows a selection marker to be gained, then lost, so that it can be used again.
In a new research paper1 from the Mitchell lab, second-year Ph.D. student Manning Huang presents a high-speed next-gen marker recycling approach called CRISPR-Cas9-Induced Marker Excision, or CRIME. The method relies upon the genome editing activity of the CRISPR-Cas9 nuclease, along with the cell's native genome repair pathways.
The method was deployed in the fungal pathogen Candida albicans to create strains with multiple lesions that block formation of infectious biofilms. Because CRIME uses the versatile CRISPR-Cas9 system and evolutionarily conserved cellular machinery, it is likely that CRIME will spread quickly in the genome science research community
1. Marker Recycling in Candida albicans through CRISPR-Cas9-Induced Marker Excision. Manning Y. Huang, Aaron P. Mitchell DOI: 10.1128/mSphere.00050-17

