Carnegie Mellon Scientists Develop High-Throughput Fluorescence Technique for Synapse Analysis
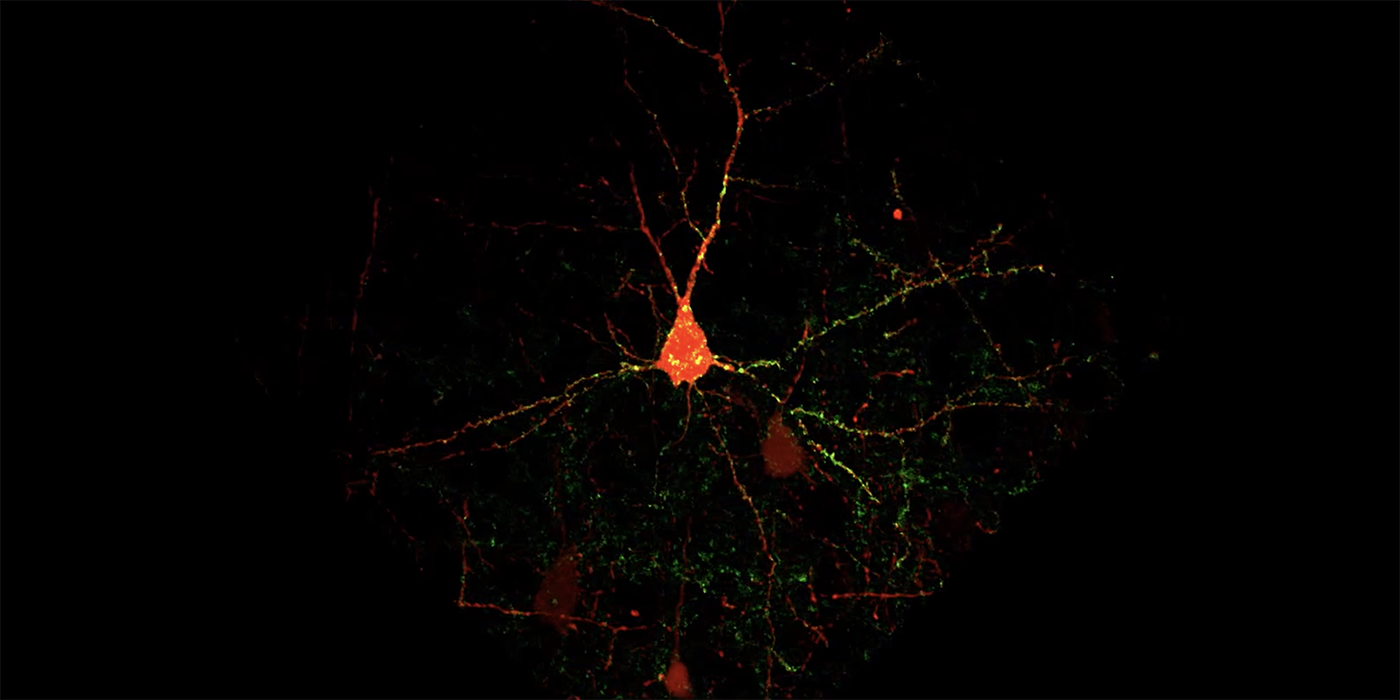
Researchers at Carnegie Mellon University’s Department of Biological Sciences have developed a new fluorescence-based technique that allows for the analysis of hundreds of thousands of synapses at a time. The new, high-throughput method for mapping the distribution of synapses on a neuron will accelerate neuroscience research on synaptic connectivity and how it changes during development, learning and disease.
To truly understand the computational properties of the neural circuits in our brain, we need to understand how and where the neurons in our brain are connected and how they transfer information between one another. But there are more than a trillion synapses in the brain, differentiated from each other by their pre- and post-synaptic partners. Counting these synapses and differentiating them from one another has been a major analytical challenge.
Traditional methods for studying synapses have not been effective to capture cell-type-specific synapses in large volumes of densely connected brain tissue. For example, electron microscopy only allows researchers to look at one part of one cell. Also, fluorescence-based antibody methods to label synapses illuminate all of the synapses, which makes it difficult to identify and quantitate specific types of synapses with molecular precision.
“It’s like analyzing the text messages for a group of people — if I was just looking at the entirety of the messages, it wouldn’t mean that much to me. But if I know who the messages were coming from, who they were going to and when they were being sent, I’d know so much more about what was going on,” said Alison Barth, the Maxwell H. and Gloria C. Connan Professor in the Life Sciences and member of the Carnegie Mellon Neuroscience Institute. “We need to have quantitative information about synapse number, dendritic location, size and input identity in order to understand the role neural circuits play in disease.”
In a study published in eNeuro, Barth and chemical biologist Marcel Bruchez devised molecular genetic methods for cell-type selective synapse labeling using the fluorogen-activating proteins (FAPs), a fluorescent probe invented and developed at Carnegie Mellon’s Molecular Biosensor and Imaging Center. Unlike traditional fluorescent proteins, FAPs only emit light when they’re bound to a fluorogen and they can be labeled with many different colors, which allows researchers to better image and differentiate between cell types.
The new technique didn’t just make imaging synapses easier, it also gave such a clear picture of synapses and their inputs that the researchers were able to create an automated computational method that counts every connection associated with a neuron, making data analysis faster and easier.
“Previous methods limited us to studying one piece of one neuron, or maybe one to ten neurons. Now we can study tens or even hundreds,” said Barth. The study, led by postdoctoral fellow Dika Kuljis, painstakingly validated the reagents and then used them to quantitatively test hypotheses about how inhibitory synapses were scattered across neocortical neurons.
“Our method gives us good, qualitative data and that reveals how different types of inhibition are distributed across the dendrites of neocortical neurons,” said Barth.
More importantly, the reagents can be applied to look at changes in synapse organization for many different cell types and disease using readily available imaging and analysis technologies.
“The barrier to use is very low for these new tools,” said Barth. “We hope that they can be broadly adopted by the neuroscience community.”
Barth’s lab is currently using their new method to study synapse function and organization in Alzheimer’s and Parkinson’s disease models as well as normal learning.
In addition to Barth, Bruchez and Kuljis, study authors include: Eunsol Park, Cheryl Telmer, Jiseok Lee and Daniel Ackerman.
This research was made possible by a grant from the Kaufman Foundation and supported by grants from the National Institute of Health (NS0902019, MH114103, NS086749).