
Imaging Technologies to Study Living Cells in Real Time
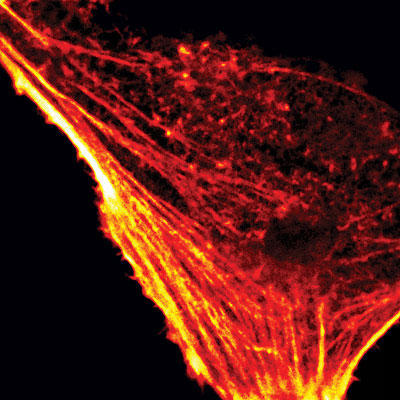
Fluorescence confocal microscope image of the actin cytoskeleton in a living HeLa cell visualized with intracellularly expressed Fluorogen Activating Protein and fluorogenic dye Malachite Green-Ester.
Trying to find one cell out of trillions in the human body or one tiny protein among the thousands inside a single cell is much like trying to pick out one star in a night sky full of billions. While astronomers build larger and larger telescopes to look deeper into space to distinguish that one celestial body of interest, scientists at the Mellon College of Science are developing smaller and smaller molecular technologies to peer more closely into living things, visualizing biological processes with greater precision and clarity than ever before.
Finding a protein needle in a cellular haystack
As scientists decode the human genome, they are gaining a better understanding of the structure of individual proteins that control the health of cells and the normal behavior of cell division. What scientists don’t know in detail is how all of the proteins interact with each other in real time in the 3D space of the cell. Uncovering these interactions could reveal how normal cells function and how things go awry in diseases like cancer and Alzheimer’s. Scientists at the Molecular Biosensor and Imaging Center (MBIC) are creating tools to do just that.
“We’re developing a powerful toolbox of intracellular fluorescent probes that will enable us to find out when a protein is being made, when it is being degraded and when it is interacting with another target protein in living cells, getting us closer to fully understanding cell networks and how they function in health and disease,” explains Alan Waggoner, professor of biological sciences and director of MBIC, a National Institutes of Health Technology Center for Networks and Pathways.
The MBIC team has created fluorogen activating proteins (FAPs), probes that emit fluorescent light only when bound to a fluorogen, an otherwise non-fluorescent dye added to the cell by the scientists. This is a step away from traditional fluorescent proteins, like green fluorescent protein (GFP), that are always aglow once expressed in cells. With the novel FAPs and associated fluorogens, the MBIC team can control when the protein lights up, a feature that is key to overcoming the difficulties inherent in pinpointing multiple, closely associated proteins inside living cells.
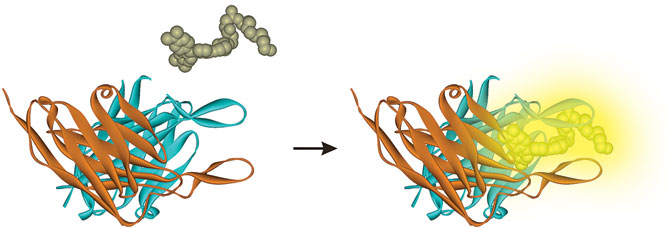
Fluorogen activating proteins (FAPs) emit fluorescent light only when bound to a fluorogen, an otherwise non-fluorescent dye added to the cell by scientists. This feature allows biologists to track proteins on the cell surface and within living cells in very simple and direct ways, eliminating cumbersome experimental steps.

Associate Research Professor of Chemistry Marcel Bruchez manages the Molecular Biosensor and Imag- ing Center’s Technology Center for Networks and Pathways, which is developing fluorescent probe and imaging technologies to investigate regulatory path- ways and networks in real-time in living cells.
“Molecules that are close together in a living cell can’t be discriminated,” explains Marcel Bruchez, associate research professor of chemistry and program manager of MBIC’s Technology Center for Networks and Pathways. “That’s just the physics of the diffraction of light.”
Imagine trying to spot five proteins that are located in the same area of the cell. If each protein were labeled with GFP, the fluorescent signals from each protein would blend together, so that all you would see through the microscope is a giant green blur. If you labeled each protein with a differently colored fluorescent probe, you would improve your ability to resolve these molecules, but there aren’t enough colors available to distinctly label all of the different proteins in a cell. However, if you labeled each of the five proteins with a FAP, you could “turn on” the fluorescence of the proteins one at a time—avoiding fluorescent interference from nearby proteins—to see exactly where in the cell each protein is located. This is possible because the two components—FAP and fluorogen—must combine to give off a signal.
“This is a very powerful method because it generates single molecule information that you can use to build a picture of the structure of the biological process or object at which you’re looking,” said Bruchez. “This new approach has the potential to do multi-color super-resolution imaging with a substantially reduced experimental complexity.”
The new FAP technology has already provided a substantial improvement in resolution, according to Bruchez. Using the FAPs, the MBIC researchers have been able to image proteins with a 1–2 nanometer accuracy. With GFP, the accuracy is approximately 20 nanometers. The beauty of MBIC’s new technology comes down to some elegant organic chemistry and molecular biology. They have created FAPs that have the properties of organic dyes, like high brightness and photostability, but that also can be genetically expressed in a cell along with a protein of interest, giving FAPs a clear advantage over current fluorescent technologies, which offer either high brightness or genetic control, but not both.
MBIC scientists are exploiting the unique properties of the FAPs to help cell biologists tackle research questions that cannot be answered using current technologies. Raymond Frizzell, professor and chair of the Department of Cell Biology and Physiology at the University of Pittsburgh, has been studying the anion channel CFTR, which spans the membrane of cells found in tissues that produce mucus. A mutation in the gene causes cystic fibrosis. Frizzell wants to know how the miscreant CFTR protein migrates to the cell surface in diseased cells compared to the normal protein in healthy cells, something that’s not possible to monitor with current technologies. The new FAP technology is already making headway in resolving this problem.
MBIC is also teaming with Diane Lidke, assistant professor in the Department of Pathology, University of New Mexico School of Medicine, to study aspects of immunity and inflammation. Consider an allergic reaction or an asthma attack. The activation of the inflammatory response happens very fast, and scientists want to know precisely how immune cells mount a defense so quickly in response to signals from their environment. Specifically, Lidke and the MBIC team are investigating Fc-epsilon receptors, which are found on certain types of white blood cells that play a major role in controlling allergic responses and promoting inflammation. They are looking at the receptor and its very early partners at the cell surface. FAPs could reveal these early events in the signaling cascade, providing new therapeutic targets that could prevent the onset of an asthma attack.
The nature of the FAPs also makes them ideal for use as biosensors, which are tools that can “sense” when a certain cellular activity, such as protein degradation, is taking place. Peter Berget, associate professor of biological sciences and a member of MBIC, has designed a FAP biosensor that responds to proteases, enzymes that cut apart proteins. Cells contain hundreds of different proteases that play important roles in everything from cancer cell metastasis to wound healing. Berget has developed a way to molecularly engineer FAPs so that they only become fluorescent when they are “cut” by a specific protease, giving off light only when the target protease is present.
Tracking cell therapies using MRI
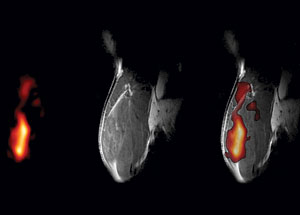
Images of a mouse quadriceps captured using a new MRI technology conceived of and developed by Eric Ahrens. (A)19F MRI image of PFPE-labeled dendritic cells. (B) Conven- tional 1H MRI image. (C) A composite 19F /1H image that places the labeled cells in their anatomical context.
Just as cell biologists strive to track protein interactions inside cells, scientists are also attempting to track the travels of individual cells among the 100 trillion cells in the human body. While spotting one individual cell might seem impossible, doing so could advance diagnostic tools and cell-based therapies including stem cell therapies and cancer vaccines.
“We now have the ability to visualize non-invasively and with sensitivity individual cells and their movement to targeted sites,” said Chien Ho, professor of biological sciences and director of the Pittsburgh NMR Center for Biomedical Research, which has been funded continuously by the National Institutes of Health since 1988. “Our new approach offers almost unlimited potential for tracking cellular and developmental processes and for monitoring cell therapies.”
According to Eric Ahrens, associate professor of biological sciences and a member of the NMR Center, many cell therapy-based clinical trials are on hold until researchers adopt a method for evaluating where the therapeutic cells go once they are administered to a patient. A new MRI technology conceived of and developed by Ahrens surmounts this obstacle by allowing researchers to safely and easily monitor the migration of such cells using MRI. Ahrens and colleagues from the University of Pittsburgh Cancer Institute are working toward using his MRI technology to monitor the migration of a cellular immunotherapy in patients with colorectal cancer. The therapy, called a dendritic cell vaccine, involves a scientist taking cells from the patient’s own body, modifying the cells to produce a tumor antigen that fights cancer cells, and reinjecting them into the patient. Ideally, the cells migrate to the lymph nodes, activating an immune response. Until now, there has been no way to know if the vaccine had reached its target unless the patient began to show improvement. Ahrens’s technique provides visual evidence that the cells have gotten to where they need to be, a requirement for FDA approval of such technologies.
Ahrens’s method involves labeling dendritic cells with a colloidal suspension of tiny fluorocarbon droplets called a perfluoropolyether (PFPE) nanoemulsion. Then, when the labeled dendritic cells are administered to the patient, they can be tracked using 19F MRI. Unlike conventional MRI, which detects the nuclear magnetic resonance signal from protons contained in the mobile water in tissue, 19F MRI detects the signal from the nucleus of the fluorine atom. Fluorine is not normally present in the body at sufficient concentrations to detect, so the PFPE-labeled cells stand out in an MRI image, making it easy to see where the vaccine has traveled.
“Dendritic cells are also being investigated for treating other types of cancers like melanoma and prostate cancer,” said Ahrens. “We’re hopeful that we can use our technology for other types of cancers in the future.”
Imaging immune responses
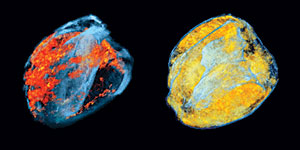
Chien Ho and colleagues in the Pittsburgh NMR Center for Biomedical Research use MRI to monitor a transplanted rat heart. These 3D renderings of MRI images show the distribution of macrophages infiltrating the heart at different stages of rejection (orange and yellow colors show the location of infiltrating macrophages).
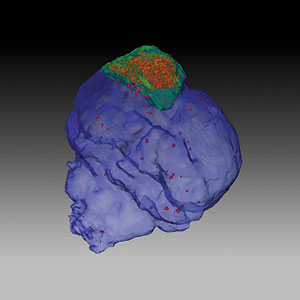
Chien Ho and colleagues in the Pittsburgh NMR Center for Biomedical Research use MRI to image a mouse brain after trauma. In this 3D rendering, the contusion is outlined in green; the MPIO-labeled macrophages appear red.

Kevin Hitchens, manager of the Pittsburgh NMR Center for Biomedical Research, runs an experiment on the Bruker AVANCE NMR scanner equipped with a 7.0 Tesla magnet. The Pittsburgh NMR Center houses a wide variety of instrumentation for Magnetic Resonance Microscopy and Magnetic Resonance Imaging that is available for use by the greater scientific community.
Dendritic cells aren’t the only immune cells NMR Center scientists are tracking by MRI. Ho and his colleagues are using MRI to track macrophages as they infiltrate a transplanted heart in the early stages of organ rejection. The real-time tracking method can pinpoint exactly when and where rejection is taking place, and it’s all done non-invasively.
Physicians typically monitor patients for organ rejection following a heart transplant by performing frequent heart biopsies, invasive procedures that involve snipping out several tiny pieces of heart tissue to look for signs of rejection. But these procedures are problematic because they only sample small areas, possibly missing the first signs of rejection. Using MRI, Ho can scan the entire heart for the presence of macrophages, immune cells that recognize the transplanted heart as foreign and set in motion an immune response that could lead to graft loss and patient death.
“Successful translation of this work to the clinic ultimately will reduce the number of biopsy procedures and should greatly improve the quality of life for cardiac transplant patients,” said Ho. “Perhaps most importantly, this advance will allow doctors to tailor the dose of immunosuppressant drugs, providing highly personalized care.”
Ho’s method involves tagging macrophages with micrometer (MPIO)-sized paramagnetic iron oxide particles, which are very sensitive to the magnetic fields used during MRI. Because a macrophage’s job is to engulf foreign materials inside the body, such as bacteria, they will also engulf MPIO particles, rendering the macrophages magnetic. After injecting MPIO particles into rats that had received heart transplants three days earlier, the researchers used MRI to track macrophages that incorporated the particles. They spotted them in transplanted hearts. The locations of the tagged macrophages are highly defined on the MRI scan, indicating that the new tracking method is very good at pinpointing where rejection is taking place.
Recently, Ho and his colleagues have applied the same technique to cases of traumatic brain injury. The inflammatory process that occurs after a traumatic brain injury can be both helpful and hurtful, repairing injured tissue but releasing neurotoxic substances that cause additional brain damage. Using their method for tagging macrophages, the scientists tracked the cells as they infiltrated the brain after injury. Because macrophages may play a pivotal role at the interface between early detrimental and delayed beneficial effects of inflammation, understanding macrophages’ role in the inflammation process could be essential to developing effective therapies for the treatment of traumatic brain injury, according to Ho.
MCS scientists continue to push the boundaries of microscopy and MRI technology, developing state of the art imaging technologies to elucidate mechanisms underlying disease processes and to track—in real time—cellular behavior in health and disease.
These advances have the potential to unlock the mysteries of the human body, much as astronomers created bigger and better telescopes to unlock the secrets of the night sky.