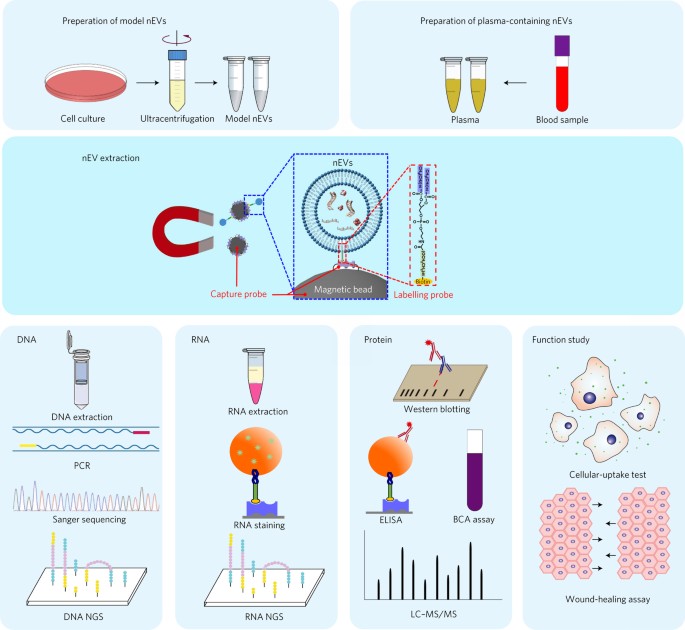
EV isolation and analysis by lipid nanoprobes
EVs can mediate intercellular communication by transferring cargo proteins and nucleic acids between cells. The pathophysiological roles and clinical value of EVs are under intense investigation, yet most studies are limited by technical challenges in the isolation of EVs. We developed a nanomaterial-based method for EV isolation. The lipid nanoprobe enables spontaneous labelling and magnetic enrichment of EVs in 15 minutes, with high isolation efficiency and purity. We also show that the lipid nanoprobes, which allow for downstream analyses of nucleic acids and proteins, enabled the identification of DNA mutations following EV isolation from blood plasma from cancer patients. The efficiency and versatility of the lipid nanoprobe opens up opportunities in point-of-care cancer diagnostics.
References:
-
Rapid isolation of extracellular vesicles via lipid nanoprobes, Y. Wan, G. Cheng, X. Liu, S.-J. Hao, M. Nisic, C.-D. Zhu, Y.-Q.Xia, W.-Q. Li, Z.-G. Wang, W.-L. Zhang, S. J. Rice, A. Sebastian, I. Albert, C. P. Belani, S.-Y. Zheng, Nature Biomedical Engineering, 1, 0058, 2017. (Feature as front cover story) [Link] [Behind the Paper] [PSU News Release]
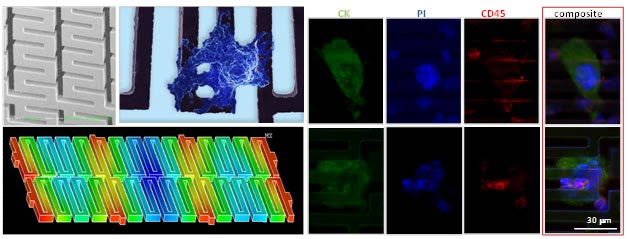
Flexible micro spring array (FMSA) for viable circulating tumor cell (CTC) enrichment
We demonstrated a high throughput versatile platform capable of isolating circulating tumor cells (CTCs) from clinically relevant volumes of blood while preserving their viability and ability to proliferate. The enrichment is based on the fact that CTCs are larger compared with normal blood cells. The incorporated system allows size-based separation of CTCs at the micro-scale, while taking advantage of a high throughput and rapid processing speed. Testing results of model systems using cell lines show that this device can enrich CTCs from 7.5 mL of whole blood samples with 90% capture efficiency, higher than 104 enrichment, and better than 80% viability in approximately ten minutes without any incidence of clogging.
References:
-
Size-based separation methods of circulating tumor cells, S.-J. Hao, Y. Wan, Y.-Q. Xia, X. Zou, and S.-Y. Zheng, Advanced Drug Delivery Reviews, 25, 3-20, 2018. [Link]
-
Evaluating a novel dimensional reduction approach for mechanical fractionation of cells using a tandem flexible micro spring array (tFMSA), Y. T. Yeh, R. A. Harouaka, S. -Y. Zheng, Lab on a Chip, 17 (4), 691-701, 2017. [Link]
-
Flexible micro spring array device for high throughput enrichment of viable circulating tumor cells, R. A. Harouaka, M.-D. Zhou, Y.-T. Yeh, W. J. Khan, A. Das, X. Liu, C. C. Christ, D. T. Dicker, T. S. Baney, J. T. Kaifi, C. P. Belani, C. I. Truica, W. S. El-Deiry, J. P. Allerton, and S.-Y. Zheng, Clinical Chemistry, vol. 60, pp. 323-333, 2014. [pdf]
-
Separable bilayer microfiltration device for viable label-free enrichment of circulating tumour cells, M.-D. Zhou†, S. Hao†, A. J. Williams†, R. A. Harouaka, B. Schrand, S. Rawal, Z. Ao, R. Brennaman, E. Gilboa, B. Lu, S. Wang, J. Zhu, R. Datar, R. Cote, Y.-C. Tai*, and S.-Y. Zheng*, Scientific Reports, 4: 7392, 1-10, doi:10.1038/srep07392, 2014. (†Authors with equal contribution) [pdf]
-
3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood, S. Zheng, H. K. Lin, B. Lu, A. Williams, R. H. Datar, R. J. Cote RJ, Y. –C. Tai, Biomedical Microdevices, vol.13, pp. 203-213, 2011. [Link]
-
Portable filter-based microdevice for detection and characterization of circulating tumor cells, H. K. Lin*, S. Zheng*, A. J. Williams, M. Balic, S. Groshen, H. I. Scher, M. Fleisher, W. Stadler, R. H. Datar, Y.-C. Tai, R. J. Cote, Clinical Cancer Research, vol 16, pp. 5011-5018, 2010. (*Authors with equal contribution)
-
Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells, S. Zheng*, H. Lin*, J.-Q. Liu, M. Balic, R. Datar, R. J. Cote, and Y.-C. Tai, Journal of Chromatography A, vol. 1162, pp. 154-161, 2007. (*Authors with equal contribution)