Biological Physics
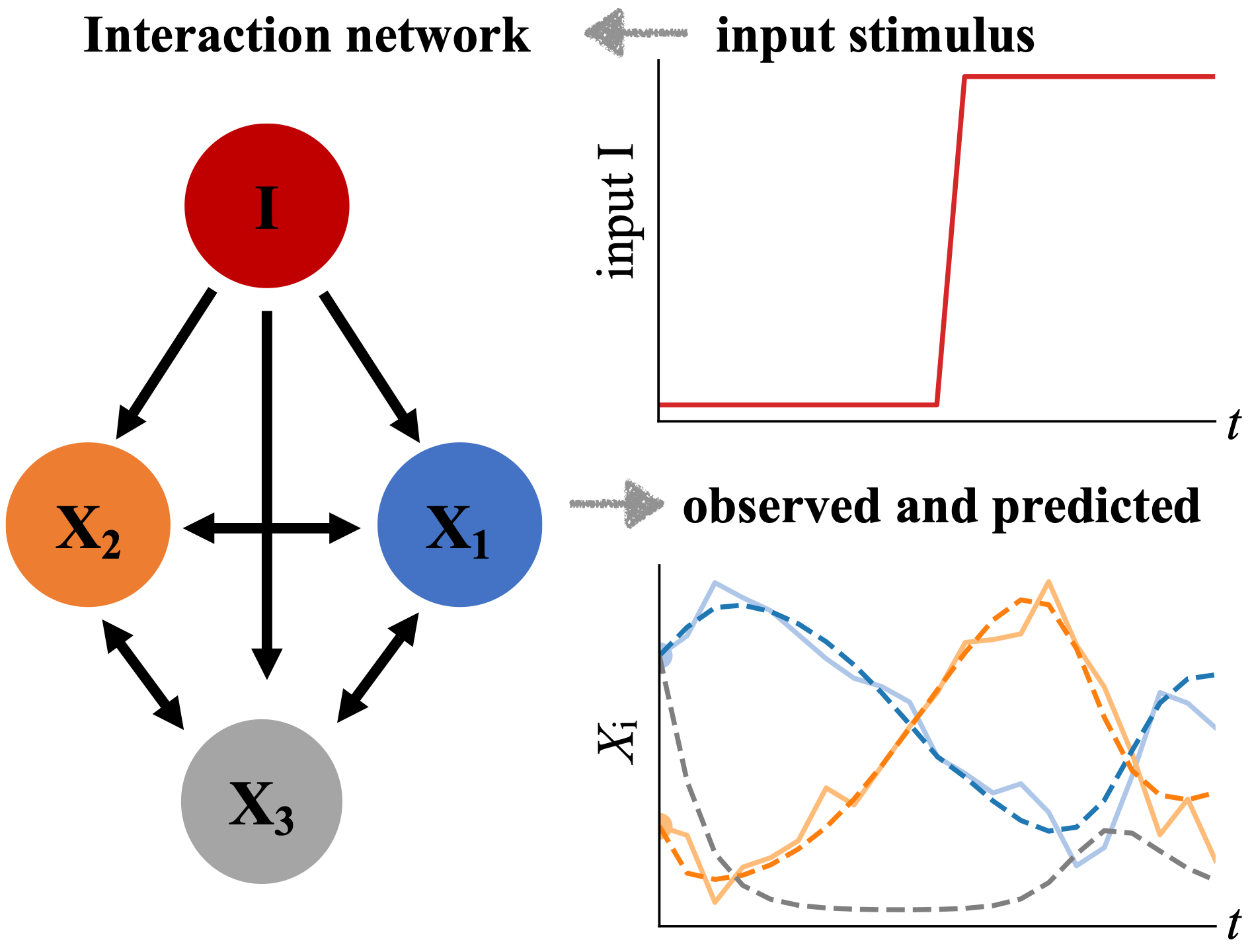
The Banerjee group is applying machine learning and inference tools to generate predictive mathematical models for the mechanical behaviors of biological cells and tissues. Their goal is to quantitatively define the regulatory interactions in living systems from time series data of experimentally observed variables, and to understand how living systems respond and adapt to environmental changes.
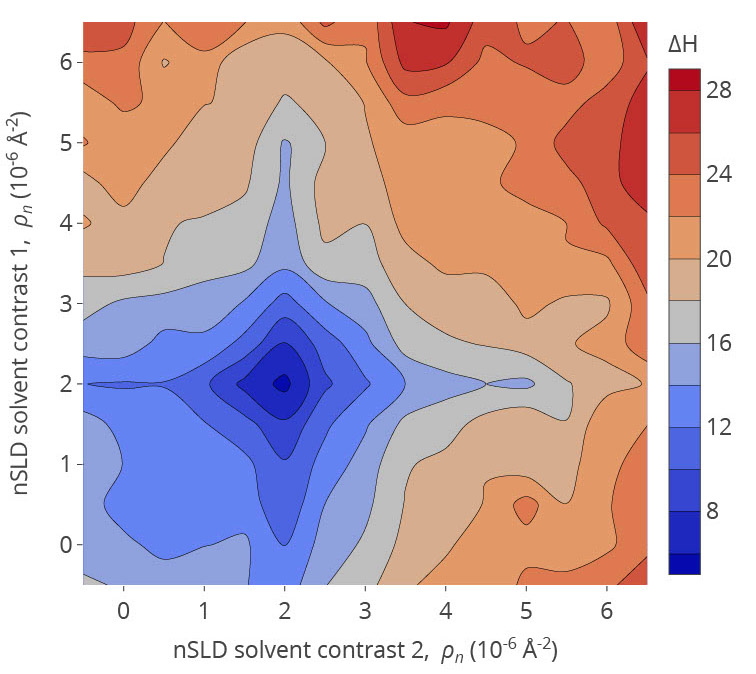
Frank Heinrich uses information theory, Bayesian statistics, and machine learning approaches to optimize neutron scattering experiments that determine the structure of lipid membrane-bound proteins. These methods are implemented primarily for instrumentation at the National Institute of Standards and Technology Center for Neutron Research. In addition, he develops an autonomous experimentation infrastructure for biological and soft matter neutron reflectometry. The image shows the information content of neutron reflectometry data (in bits, higher is better) from a lipid membrane as a function of the aqueous bulk solvent's tuneable isotopic composition (nSLD) in two successive measurements.
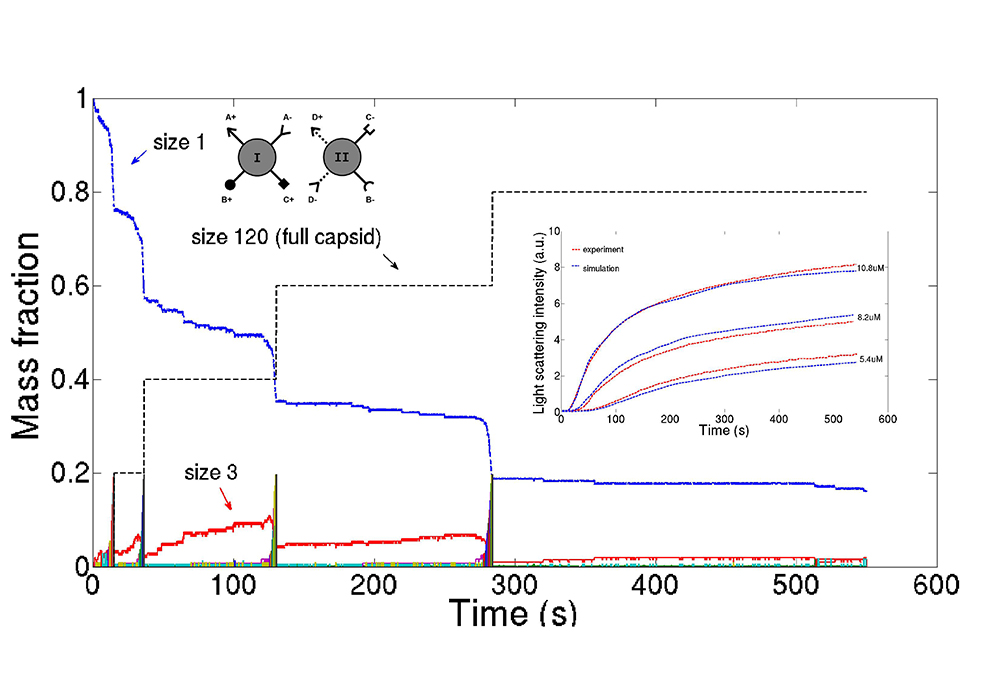
The Schwartz lab is developing methods using AI, machine learning, and optimization algorithms to learn such models from experimental data to allow us to study features of the assembly process that are crucial to their function but that cannot be directly observed experimentally. This image shows time progress of a stochastic computer simulation of viral assembly based on a model learned through simulation-based data fitting.