NEWS & Events
- Graduate Student Ting Liu successfully defended her PhD thesis on Apr.29th, 2016. Thesis title: "Biophysics of encapsidated DNA states in viruses and their role for infectivity".
- Aug 20, 2015, graduate student Ting Liu selected to the inaugural class of CMU Presidential Fellows as a recipient of Bruce McWilliams Presidential Fellowship in the Mellon College of Science 2015-16.
- July 20, 2015, Carnegie Mellon University Press Release "Scientists identify achilles' heel of virus's tough outer shell."
- Graduate Student Dave Bauer successfully defended his phD thesis on Apr.13th, 2015. Thesis title: "Influence of internal genome pressure on viral particle infectivity and stability".
- "Graduate Student Krista Freeman is selected to attend the 65th Lindau Nobel Laureate Meeting in Lindau, Germany." Only the 55 most qualified young scientists from the United States are given the opportunity to enrich and share the unique atmosphere of the Lindau Nobel Laureate Meetings.The meeting is held from 28 June to 3 July 2015.
- "Rapid transitions in viral DNA story, highlighted in PNAS In this issue front page" PNAS, October 14, 2014
- "Body temperature linked to DNA activity inside disease-causing virus" Pittsburgh Post-Gazette, November 11, 2014
- September 30, 2014, Carnegie Mellon University Press Release "Viral Infection Might Just Be a Phase Transition"
- Graduate Student Dave Bauer was awarded an NIH fellowship from MBSB graduate program based on his successful grant application.
- Listen to the popular science podcast "The Skeptics' Guide to the Universe" in the final segment, "Science or Fiction" starting around 63 min our discovery of Herpes pressure is being discussed.
- July 24, 2013, Media Press Release "First Experimental Evidence of Internal Pressure Inside Herpes Virus".
- Graduate Student Krista Freeman was awarded a 2013 NSF Graduate Research Fellowship Program (GRFP) Fellowship.
- Graduate Student Dave Bauer is awarded an Outstanding Presentation Prize at the Grad Expo 2013 at the University of Pittsburgh (March 21, 2013).
- Professor Evilevitch has been awarded an NSF grant (June 2012).
- Graduate Student Udom Sae-Ueng receives Astrid and Bruce McWilliams Fellowship for impressive research accomplishments in Physical Virology (February 2012).
- "Physicist Makes Pivotal Discovery", Carnegie Mellon Front Page, July 17th, 2010.
- "Researcher says physics may outsmart viruses" Pittsburgh Post-Gazette Wednesday, February 17th, 2010
- "Carnegie Mellon Physicist the First To Measure Energy Released From a Virus During Infection" Carnegie Mellon University press release, February 5th, 2010
- "Energy of Attacking Virus Revealed" Science Daily,
January 2010 - "High-powered living DNA cannon" Nano Tsunami - Nano Medicine in depth, 2005
- "DNA Goes Ballistic" Science,
July 2003 - "Some viruses faster than a speeding bullet" ABC News,
July 2003
Research Outline
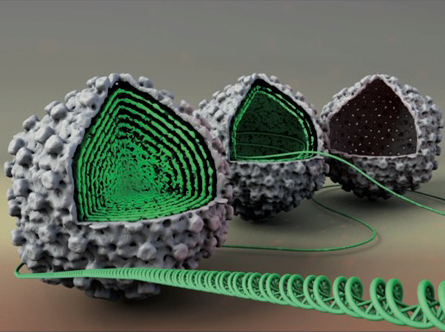
The majority of viruses possess spherical, icosahedral protein shells with radii varying between 10 nm and 100 nm and with thicknesses of a few nanometers corresponding to single protein layer. Viral capsids protect genomes that are tens of microns in contour length. Sufficient genome encapsidation implies that the virus must be stable enough to withstand internal forces exerted by its packaged genome and external forces from its environment. Yet, it must be unstable enough to rapidly release its genome in the cell during infection. Thus, there must exist a unique match between the virus’s genome length and capsid size and strength that is adjusted to the biological and physical properties of the host cell. Internal genome pressure, reaching tens of atmospheres as a result of strong confinement, is required for phages and many other dsDNA viruses to be able to infect by passive ejection of its genome. Besides from determining this pressure, we also found that it provides additional support to the strength of the viral capsid helping the virus survive external deformations imposed on it between infections.
Our group uses biophysical approaches in order to learn about the fundamental physical principles that control viral genome encapsidation and release as well as capsid stability. This research program takes advantage of the high-resolution cryo electron microscopy, AFM, light scattering and microcalorimetry. Furthermore, our findings provide tools for the rational design of therapeutic agents that selectively interfere with the encapsidation process, and in addition tools to improve encapsidation in vitro in order to make stable vectors for gene delivery.
How to interfere with success of the virus?
Success of the virus, i.e. the infectivity, depends on the match between genome length and capsid size:
1. Ability to bring viral genome in the cell
VIRAL GENOME: EJECTION AND PACKAGING
- Pressure inside Human Herpes Simplex Virus (HSV).
- Control of capsid pressure by varying electrolyte concentrations. (Ref. 16, 10, 9)
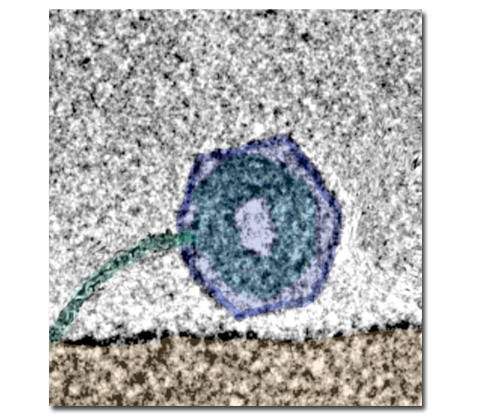
- Ion distribution in viruses studied with EFTEM.
- Direct calorimetric measurements of energy stored in the packaged DNA of viral particles. (Ref. 21)
- DNA structure inside the viral capsids.
- Dynamics of viral DNA ejection. (Ref. 13, 11)
- Microfluidics as a tool to study viral infection in vivo.
- Osmotic pressure - promoting and resisting viral DNA release (Ref. 17, 16, 14, 10, 9, 8, 7, 6, 5)
2. How to survive external mechanical capsid stress
before DNA release?
VIRAL CAPSID: MECHANICAL PROPERTIES, PACKAGING MOTOR, AND SELF-ASSEMBLY
- Effect of internal DNA pressure on viral capsid stability. (Ref. 12)
- Evolutionary optimization of viruses.
- Viral DNA packaging motor – strongest molecular motor known.