Expanding a Model Gene Regulatory Network
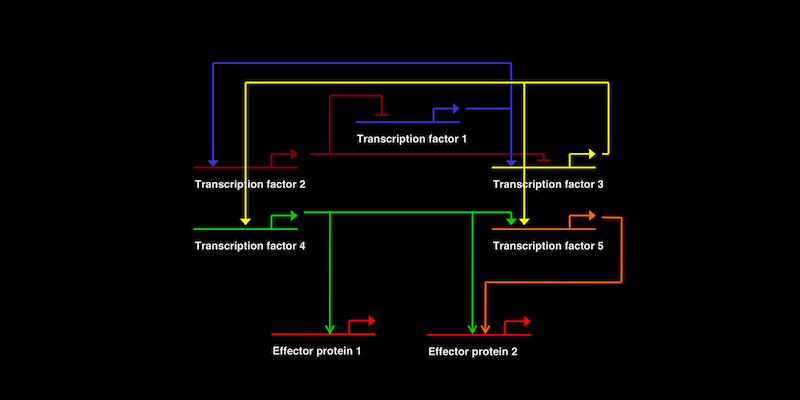
Recently, the Ettensohn lab published two papers that expand a model gene regulatory network (one that controls the formation of the skeleton in sea urchins) by identifying important new genes in this network.
The lab studies developmental gene regulatory networks – dynamic genetic networks that drive programs of differential gene expression and that endow embryonic cells with their unique identities. Sea urchins are used as a model organism because they are particularly well suited to the experimental analysis of embryonic development and gene networks.
In the papers, the lab identifies two new genes. One gene, KirrelL, encodes a cell adhesion protein that mediates cell-cell fusion- an important behavior of the embryonic cells that produce the skeleton (Ettensohn and Dey, 20171). The second gene, TgfbrtII, encodes a cell surface receptor that transduces signals from TGFβ-family ligands (Sun and Ettensohn, 20172). This protein acts later in development and mediates signals from neighboring embryonic tissues that regulate the formation of the skeleton.
Together, these two papers add to our understanding of developmental gene regulatory networks and how they control early animal development.
1. Ettensohn, C. A., & Dey, D. (2017). KirrelL, a member of the ig-domain superfamily of adhesion proteins, is essential for fusion of primary mesenchyme cells in the sea urchin embryo. Developmental Biology, 421(2), 258-270. doi:10.1016/j.ydbio.2016.11.006
2. Sun, Z., & Ettensohn, C. A. (2017). TGF-β sensu stricto signaling regulates skeletal morphogenesis in the sea urchin embryo. Developmental Biology, 421(2), 149-160. doi:10.1016/j.ydbio.2016.12.007