
Bruce Armitage
Professor of Chemistry & Co-Director of CNAST
Research
The Armitage Lab is broadly interested in the use of nucleic acids and their synthetic analogues for applications in biomedical research and biotechnology development. Much of our work relies on peptide nucleic acid (PNA. Figure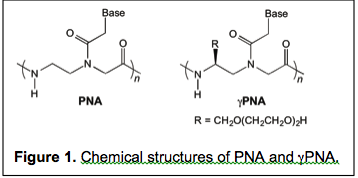 1), a DNA mimic invented at the University of Copenhagen in 1991 by Nielsen, Egholm, Berg and Buchardt. In the first two projects described below, we are using PNA to hybridize to specific DNA or RNA targets with the goals of (a) manipulating gene expression or (b) report on the presence and/or activity of a nucleic acid. The third project describes our efforts to use DNA nanotechnology to assemble bright, multi-dimensional fluorescent nanostructures for use in biological imaging and sensing.
1), a DNA mimic invented at the University of Copenhagen in 1991 by Nielsen, Egholm, Berg and Buchardt. In the first two projects described below, we are using PNA to hybridize to specific DNA or RNA targets with the goals of (a) manipulating gene expression or (b) report on the presence and/or activity of a nucleic acid. The third project describes our efforts to use DNA nanotechnology to assemble bright, multi-dimensional fluorescent nanostructures for use in biological imaging and sensing.
1. Guanine Heteroquadruplex Formation by PNA. Watson and Crick forever linked guanine with cytosine in their seminal 1953 paper proposing the double helical structure of DNA. However, research by Davies and Gellert in the 1960s led to the proposal that guanine could self assemble into G-tetrads, stabilized by hydrogen bonding and metal ion coordination. Pi-stacks of such tetrads gives rise to the higher order family of structures known as G-quadruplexes. Biological interest in quadruplex DNA and RNA has grown steadily in the past two decades, due in large part to (1) strong conservation of QFSs in the repeating telomere sequences found at the ends of chromosomes; (2) bioinformatics studies showing nonstatistical distributions of quadruplex-forming sequences in gene regulatory regions (e.g. promoters, UTRs); (3) discovery of numerous endogenous proteins with quadruplex-binding ability and (4) demonstration of functional activities in reporter assays. The mounting evidence in favor of biologically important functions for G-quadruplexes has in turn motivated chemists to design and synthesize ligands that can bind to these structures, in turn reporting on their presence or manipulating their activity.
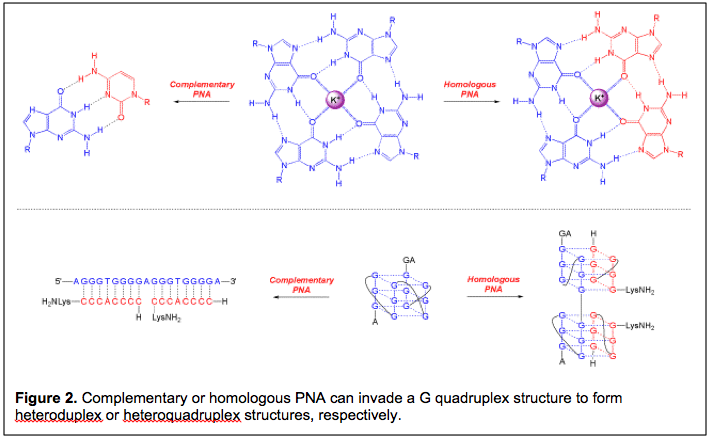
Our group entered the area of quadruplex recognition in 2001, with a paper that described the ability of cytosine-rich PNA to hybridize to a complementary DNA target embedded within a folded G-quadruplex structure. (Figure 2; subsequent work extended this phenomenon to RNA quadruplexes.) These were examples of classical DNA/RNA targeting using the simple rules laid out by Watson and Crick: G pairs with C and A pairs with T. The high affinity of PNA allowed it to invade the folded quadruplex structures in order to form PNA-DNA or PNA-RNA heteroduplexes.
In addition to targeting G-quadruplexes using complementary PNA oligomers, a former student in the lab, Bhaskar Datta, proposed and demonstrated that homologous PNA oligomers could also bind to DNA G-quadruplexes. In these cases, the resulting hybrid is a heteroquadruplex in which each G tetrad is comprised of guanines from both the DNA target and the PNA ligand. Subsequent work demonstrated that the affinities of homologous PNAs for their G quadruplex targets are usually in the low nanomolar range. After a series of studies reporting on the thermodynamics and kinetics of heteroquadruplex formation as well as improvements to the PNA design to improve selectivity, we are now embarking on a program to study the functional properties of quadruplex-forming PNAs, relying first on gene expression reporter assays with subsequent efforts to focus on assessing impacts of these PNAs on endogenous gene expression. The ultimate goal of this chemical biology project is to use quadruplex-forming PNAs to regulate expression of specific genes at the transcriptional and/or translational levels.
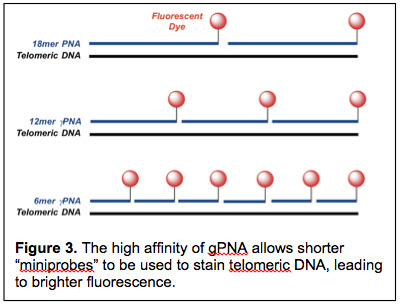
The most common method for measuring telomere length involves a fluorescent PNA probe consisting of the sequence (CCCAAT)3 (Figure 3). This PNA telomere FISH probe is complementary to the corresponding repeating sequence of human telomeres, i.e. 5’-(GGGTTA)n-3’, and is widely used for quantitative length determinations (Q-FISH). However, a challenge with using this probe for telomere analysis arises from low signal from short telomeres. As humans age, their telomeres become progressive shorter and there is growing awareness that critically short telomeres, i.e. telomeres that have shortened below a certain length, may be more important in determining susceptibility to certain diseases than is the average length of all telomeres. The detection problem arises from the fact that, as a telomere shortens, fewer PNA FISH probes can bind, leading to lower fluorescence intensity.
In collaboration with CNAST member Patty Opresko, we have been developing higher affinity FISH probes for telomere analysis. Using the γPNA modifications invented by Danith Ly, we have been able to shorten the FISH probe from the standard 18mer to 12mer and 6mer versions that retain the ability to bind and fluorescently label telomeric DNA. The shorter length of these miniprobes translates into binding of more probes/telomere, ultimately resulting in brighter fluorescence. Our ongoing efforts on this project are directed toward developing even brighter FISH probes through attachment of additional dyes to the γPNA. In addition, we are working on extending the miniprobe concept to RNA FISH probes through the use of libraries of short γPNA miniprobes that are targeted to adjacent sites along an RNA.
3. Fluorescent DNA Nanotags. Our third nucleic acid project relies on DNA nanotechnology to produce well-defined two- and three-dimensional DNA objects. The key to DNA nanotech is to design DNA strands that are not completely complementary to one another. For example, if three DNA strands are designed such that each strand is partially complementary to the other two, then the three strands can assemble into a three-way junction, i.e. a 2-D structure. Alternatively, if four DNA strands are designed such that each strand is partially complementary to the other three, then a 3-D tetrahedron can be prepared (e.g. Figure 4).
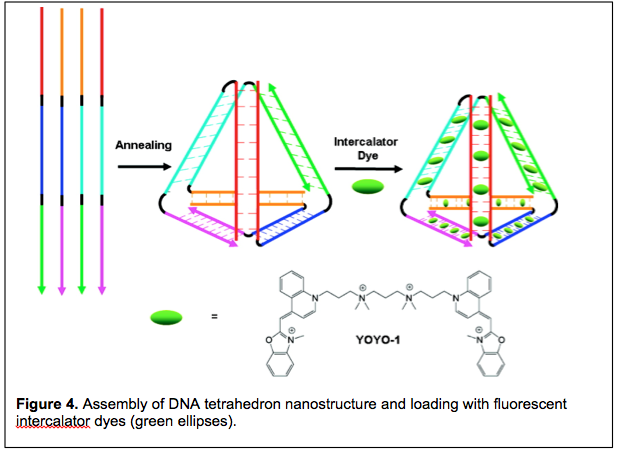
We are exploiting DNA nanotechnology to create a new class of fluorescent labels that promise to be much brighter than existing fluorescent dyes. The idea takes advantage of the well-known intercalation mode by which planar dye molecules bind to DNA via insertion between adjacent base pairs. Intercalation can occur up to a density of one dye for every two base pairs. Hence, for a nanostructure having 100 base pairs, up to 50 dye molecules can be intercalated. Since the brightness of a fluorophore is defined as the product of the extinction coefficient (i.e. the efficiency of light absorption) and the quantum yield (i.e. the efficiency of light emission), dye-loaded DNA nanostructures have the potential to be exceptionally bright fluorescent objects. When used to fluorescently label a molecule or structure of interest, these assemblies are referred to as fluorescent DNA nanotags.
Published work from our lab describes the initial design and characterization of 2-D and 3-D nanotags and their use in fluorescent labeling of cell surfaces and polymer beads. Incorporation of longer wavelength dyes leads to exceptionally efficient energy transfer, resulting in shifting of the fluorescence to longer wavelength and minimizing background signal. Ongoing work is directed toward enhancing the stability of fluorescent nanotags to allow them to be used to label intracellular targets. We collaborate with the Peteanu, Lopez and McCartney labs on subprojects ranging from single molecule imaging and analysis to RNA and protein labeling.
Publications
Gupta, A.; Lee, L.-L.; Roy, S.; Tanious, F. A.; Wilson, W. D.; Ly, D. H.; Armitage, B. A. Strand Invasion of DNA Quadruplexes by PNA: Comparison of Homologous and Complementary Hybridization. ChemBioChem 2013 In press.Goldman, J. M.; Zhang, L. A.; Manna, A.; Armitage, B. A.; Ly, D. H.; Schneider, J. W. High Affinity γPNA Sandwich Hybridization Assay for Rapid Detection of Short Nucleic Acid Targets with Single Mismatch Discrimination. Biomacromolecules 2013 In press.
Thomas, S. M.; Sahu, B.; Rapireddy, S.; Bahal, R.; Wheeler, S. E.; Procopio, E. M.; Kim, J.; Joyce, S. C.; Contrucci, S.; Wang, Y.; Chiosea, S. I.; Lathrop, K. L.; Watkins, S.; Grandis, J. R.; Armitage, B. A.; Ly, D. H. Antitumor Effects of EGFR Antisense Guanidine-Based Peptide Nucleic Acids in Cancer Models. ACS Chem. Biol. 2013, 8, 345-352. PMID: 23113581.
Stadler, A. L.; Delos Santos, J. O.; Stensrud, E. S.; Dembska, A.; Silva, G. L.; Liu, S.; Shank, N. I.; Kunttas-Tatli, E.; Sobers, C. J.; Gramlich, P. M. E.; Carell, T.; Peteanu, L. A.; McCartney, B. M.; Armitage, B. A. Fluorescent DNA Nanotags Featuring Covalently Attached Intercalating Dyes: Synthesis, Antibody Conjugation and Intracellular Imaging. Bioconjugate Chem. 2011, 22, 1491-1502. PMID: 21755981.
Mohammed, H. S.; Delos Santos, J. O.; Armitage, B. A. Noncovalent Binding and Fluorogenic Response of Cyanine Dyes to DNA Homoquadruplex and PNAD-DNA Heteroquadruplex Structures. Artif. DNA PNA XNA 2011, 2, 43-49. PMID: 21912726.
Roy, S.; Tanious, F. A.; Zanotti, K. J.; Murphy, C. T.; Wilson, W. D.; Ly, D. H.; Armitage, B. A. Kinetic Discrimination of DNA G Quadruplex Targets by Guanine Rich Peptide Nucleic Acids. Chem. Commun. 2011, 47, 8524-8526.
Sahu, B.; Sacui, I.; Rapireddy, S.; Zanotti, K. J.; Bahal, R.; Armitage, B. A.; Ly, D. H. Synthesis and Characterization of Conformationally Preorganized, (R)-Diethylene Glycol-Containing γ-Peptide Nucleic Acids with Superior Hybridization Properties and Water Solubility. J. Org. Chem. 2011, 76, 5614-5627.
Lusvarghi, S.; Murphy, C. T.; Roy, S.; Tanious, F. A.; Sacui, I.; Wilson, W. D.; Ly, D. H.; Armitage, B. A. Loop and Backbone Modifications of PNA Improve G Quadruplex Binding Selectivity. J. Am. Chem. Soc. 2009, 131, 18415-18424.
Özhalici-Ünal, H.; Armitage, B. A. Fluorescent DNA Nanotags Based on a Self-Assembled DNA Tetrahedron. ACS Nano 2009, 3, 425-433.
Constantin, T. P.; Silva, G. L.; Robertson, K. L.; Hamilton, T. P.; Fague, K.; Waggoner, A. S.; Armitage, B. A. Synthesis of New Fluorogenic Cyanine Dyes and Incorporation into RNA Fluoromodules. Org. Lett. 2008, 10, 1561-1564.