| |
|
|
Membrane Association of α-Hemolysin:
Proteins Functionally Reconstituted in tBLMs
Duncan J. McGillivray,§,†,‡ Frank Heinrich,§,† Gintaras Valincius,* Ilja Ignatjev,* David J. Vanderah,† Mathias Lösche§,†, and John J. Kasianowicz†
§Carnegie Mellon University, Pittsburgh, PA; †National Institute of Standards and Technology, Gaithersburg, MD; *Institute of Biochemistry, Vilnius, Lithuania
‡Now at the Dept. of Chemistry, The University of Auckland, New Zealand.
The protein α-hemolysin (αHL) is a pore-forming toxin from the bacterium, Staphylococcus aureus, which shows promise for development as a stochastic biosensor for small ions [1], proteins [2] and large molecules, including DNA [3,4].
We have incorporated the protein into a robust, biomimetic tethered bilayer lipid membrane (tBLM). Monitoring the reconstituted pore’s ion transport properties with electrical impedance spectroscopy we verified that the toxin retains its natural functionality. This has enabled the characterization of the interaction between the membrane and protein, using neutron reflectometry at the Advanced Neutron Reflectometer / Diffractometer (AND/R) [5]. |
Tethered Bilayer Membranes (tBLMs)
Technological challenges in the design of a robust interfacial architecture that combines a fluid bilayer with embedded proteins, for example for the design of a protein-based biosensor:
• Organic surface films should be robust, and preferably and reusable
• Protein activity must be preserved
•
Protein response must be easily measurable
 Our solution: We devised synthetic thiolipids in which a thiol is separated from a hydrophobic lipid anchor by a hydrophilic oligo(ethyleneoxide) spacer [6,7]. Self-assembled monolayers (SAMs) of these compounds are formed on gold-coated substrates and completed with phospholipids (for example, diphytanoyl-phosphatidylcholine, DPhyPC). In the completed surface-supported membrane, the lipids are decoupled from the gold surface by the spacer, as a nanometer-thin, stable aqueous layer is formed between the membrane and the substrate. Neutron reflectometry (NR) and electrical impedance spectroscopy (EIS) show complete, electrically-sealing tBLMs, that can cover large areas (cm2) and are stable for days [8]. In fact, similar constructs have been shown to be stable over months [9]. Our solution: We devised synthetic thiolipids in which a thiol is separated from a hydrophobic lipid anchor by a hydrophilic oligo(ethyleneoxide) spacer [6,7]. Self-assembled monolayers (SAMs) of these compounds are formed on gold-coated substrates and completed with phospholipids (for example, diphytanoyl-phosphatidylcholine, DPhyPC). In the completed surface-supported membrane, the lipids are decoupled from the gold surface by the spacer, as a nanometer-thin, stable aqueous layer is formed between the membrane and the substrate. Neutron reflectometry (NR) and electrical impedance spectroscopy (EIS) show complete, electrically-sealing tBLMs, that can cover large areas (cm2) and are stable for days [8]. In fact, similar constructs have been shown to be stable over months [9].
To ensure hydration of the submembrane space requires "backfilling", i.e., spacing out the grafted thiolipids in-plane with a diluent, such as 2-mercaptoethanol (βME). tBLMs can be completed with a range of lipids, saturated or unsaturated, charged or zwitterionic.
|
Protein Reconstitution and Functionality
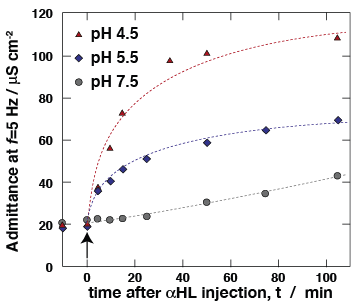 Ion transport properties of αHL, as determined in free-standing "black" lipid membranes (BLMs), are a benchmark of the protein activity. Using electrochemical impedance spectroscopy (EIS), we measure membrane conductivity of tBLMs following incubation with αHL, thereby monitoring protein reconstitution. At low pH, αHL reconstitutes more rapidly in the membrane, matching previously reported reconstitution work into freestanding BLMs [10]. Ion transport properties of αHL, as determined in free-standing "black" lipid membranes (BLMs), are a benchmark of the protein activity. Using electrochemical impedance spectroscopy (EIS), we measure membrane conductivity of tBLMs following incubation with αHL, thereby monitoring protein reconstitution. At low pH, αHL reconstitutes more rapidly in the membrane, matching previously reported reconstitution work into freestanding BLMs [10].
Membrane-reconstituted 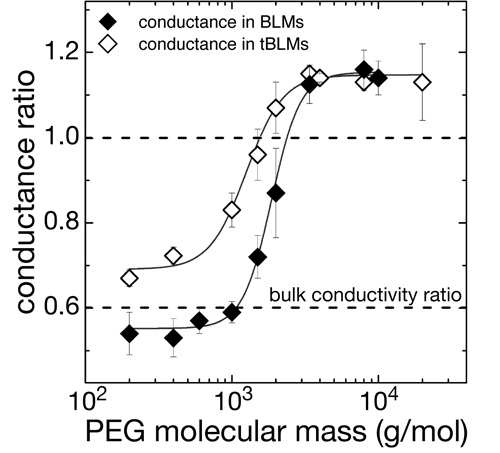 αHL pores are penetrated by water-soluble polymers such as PEG if the polymer chain length is sufficiently small [11]. PEG penetration into the pores restricts the transport of small ions through the channels, which can be used to monitor the interaction of the polymer with the pore with high sensitivity. We measure this restriction of ion conductivity in tBLMs as a function of PEG molecular weight and observe a MW-dependence similar to that observed in free-standing BLMs. Conclusion: Reconstituted αHL forms active, fully functional membrane pores in tBLMs at high lateral density. αHL pores are penetrated by water-soluble polymers such as PEG if the polymer chain length is sufficiently small [11]. PEG penetration into the pores restricts the transport of small ions through the channels, which can be used to monitor the interaction of the polymer with the pore with high sensitivity. We measure this restriction of ion conductivity in tBLMs as a function of PEG molecular weight and observe a MW-dependence similar to that observed in free-standing BLMs. Conclusion: Reconstituted αHL forms active, fully functional membrane pores in tBLMs at high lateral density.
|
Neutron Reflection Reveals Structure of a Membrane-Reconstituted Protein and its Association with the Membrane
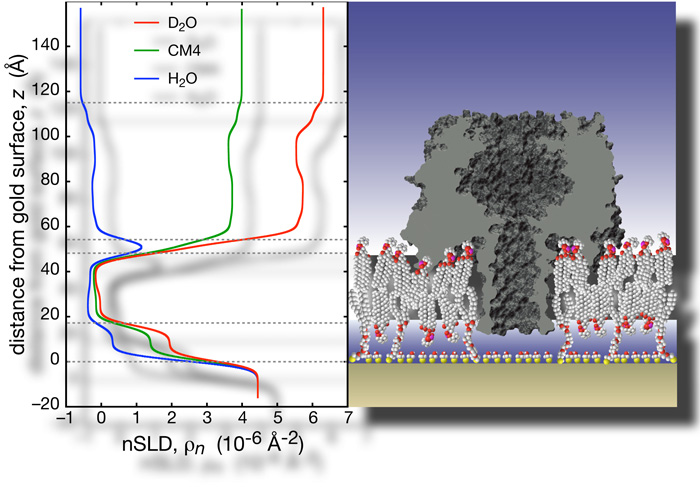
We performed extensive structural studies of αHL-reconstituted tBLMs using neutron reflection (NR) measurements. These studies were facilitated by the superior resilience of the tethered membrane, which permits multiple solvent exchanges in situ on the AND/R, followed by repeated data acquisition scans. Because these scans are all measured with identical beam footprints on the sample, we can co-refine multiple data sets with one molecular parameter set that describes the supramolecular structure. Several new data evaluation procedures were developed in the course of this work.
The heptameric pore can be modeled using the available X-ray structure [12]. The PDB coordinates were used to generate a projection of neutron scattering lengths (nSL) and protein volume occupancy along the symmetry axis. These capabilities are now freely available. All isotopically distinct data sets, 5 in total, can be simultaneously fitted using ga_refl [13]. These fits, derived from one consistent structure, describe the experimental data with high fidelity. The resolution of the structure along the direction of surface normal, i.e., the confidence interval in which the protein can be located within the membrane, is ~ 1 Ångstrom. |
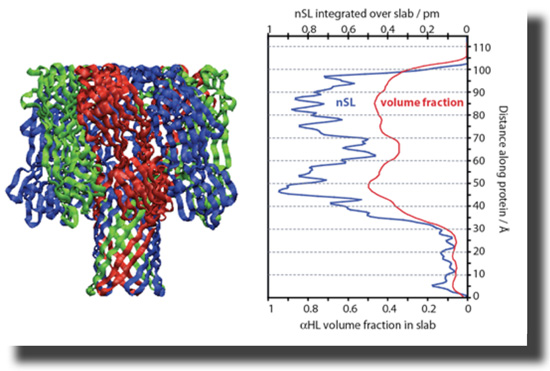
With help from Hirsh Nanda, Frank designed a script that converts PDB coordinates into nSL distributions which are appropriately averaged along the z axis. The projection of the protein on the surface normal can be tilted. The resulting nSL distribution can be shifted with respect to the bilayer to determine the depth of insertion of the protein, and thus details of the association of the protein with the membrane. |
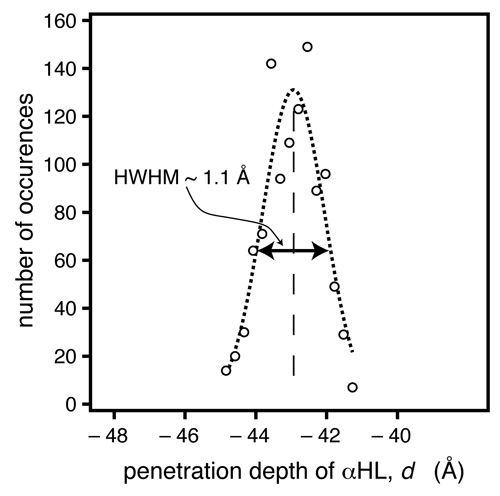 In another methodic development, Frank implemented a Monte-Carlo (MC)-based data evaluation procedure [14], in which token data sets are generated, based upon the experimental data and their associated measurement errors. When we fit these token data sets, a large number of parameter sets are determined that could have occurred given the quality of the data. Evaluation of these parameter sets, e.g., in the form of histograms, provides bias-free estimators of the parameter confidence limits [15]. In another methodic development, Frank implemented a Monte-Carlo (MC)-based data evaluation procedure [14], in which token data sets are generated, based upon the experimental data and their associated measurement errors. When we fit these token data sets, a large number of parameter sets are determined that could have occurred given the quality of the data. Evaluation of these parameter sets, e.g., in the form of histograms, provides bias-free estimators of the parameter confidence limits [15].
For the αHL system investigated here, the MC procedure indicates we can determine the location of the protein within the bilayer, as projected on the z axis, with a precision of ~ 1 Å. |
The results show that αHL interacts strongly with the bulk-solvent exposed outer headgroups, and perturbs the tail region with its rim domains. The protein stem penetrates the hydrophobic barrier, and extends ~5 Å into the headgroups of the lipid layer next to the substrate. These observations match predictions from the crystal structure, based on the locations of charged and non-polar residues. The surface coverage is estimated to be ~33% of the maximum packing density [16].
|
This research has been reported in McGillivray, D. J., et al. (2009), Biophys. J. 96, 1547, ref. [17] below. It was mainly supported by the National Science Foundation (CBET #0555201 and 0457148). This project benefited greatly from neutron research facilities provided by the National Institute of Standards and Technology at the NIST Center for Neutron Research.
References
[1] Bezrukov, S. M.; Kasianowicz, J. J. (1993), Phys. Rev. Lett. 70, 2352.
[2] Kasianowicz, J. J.; Henrickson, S. E.; Weetall, H. H.; Robertson, B. (2001), Anal. Chem. 73, 2268.
[3] Kasianowicz, J. J.; Brandin, E.; Branton, D.; Deamer, D. W. (1996), Proc. Natl. Acad. Sci. USA 93, 13770.
[4] Kasianowicz, J. J. (2004), Nat. Mater. 3, 355.
[5] Dura, J. A.; et al. (2006), Rev. Sci. Instr. 77, 074301.
[6] McGillivray, D. J.; Valincius, G.; Vanderah D. J.; Febo-Ayala, W.; Woodward, J. T.; Heinrich, F.; Kasianowicz, J. J.; Lösche, M. (2007), Biointerphases 2, 21.
[7] Heinrich F.; Ng, T.; Vanderah, D. J.; Shekhar, P.; Mihailescu, M.; Nanda, H.; Lösche, M. (2009), Langmuir 25, in press.
[8] Valincius, G.; McGillivray, D. J.; Febo-Ayala, W.; Vanderah, D. J.; Kasianowicz, J. J.; Lösche, M. (2006), J. Phys. Chem. B 110, 10213.
[9]
Vockenroth, I. K.; Ohm, C.; Robertson, J. W. F.; McGillivray, D. J.; Lösche, M.; Köper, I. (2008), Biointerphases 3, FA68.
[10] Kasianowicz, J. J.; Bezrukov, S. M. (1995), Biophys. J. 69, 94.
[11] Bezrukov, S. M.; Vodyanoy, I.; Brutyan, R. A.; Kasianowicz, J. J. (1996), Macromolecules 29, 8517.
[12] Song, L. Z.; Hobaugh, M. R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J. E. (1996), Science 274, 1859.
[13] Kienzle, P. A.; Doucet, M.; McGillivray, D. J.; O'Donovan, K. V.; Berk, N. F.; Majkrzak, C. F. (2000-2009). http://www.ncnr.nist.gov/reflpak/garefl.html.
[14] Press, W. H.; Flannery, B. P.; Teukolsky, S. A.; Vetterling, W. T. (1986). Numerical Recipes, Cambridge University Press, chapter 14.
[15] Heinrich F.; Ng, T.; Vanderah, D. J.; Shekhar, P.; Mihailescu, M.; Nanda, H.; Lösche, M. (2009), Langmuir 25, in press.
[16] McGillivray, D. J.; Valincius, G.; Heinrich, F.; Robertson, J. W. F.; Vanderah. D. J.; Febo-Ayala, W. Ignatjev, I.; Lösche, M.; Kasianowicz, J. J. (2009), Biophys. J. 96, 1547.
|
|
|
|

 In another methodic development, Frank implemented a Monte-Carlo (MC)-based data evaluation procedure [14], in which token data sets are generated, based upon the experimental data and their associated measurement errors. When we fit these token data sets, a large number of parameter sets are determined that could have occurred given the quality of the data. Evaluation of these parameter sets, e.g., in the form of histograms, provides bias-free estimators of the parameter confidence limits [15].
In another methodic development, Frank implemented a Monte-Carlo (MC)-based data evaluation procedure [14], in which token data sets are generated, based upon the experimental data and their associated measurement errors. When we fit these token data sets, a large number of parameter sets are determined that could have occurred given the quality of the data. Evaluation of these parameter sets, e.g., in the form of histograms, provides bias-free estimators of the parameter confidence limits [15].



 αHL pores are penetrated by water-soluble polymers such as PEG if the polymer chain length is sufficiently small
αHL pores are penetrated by water-soluble polymers such as PEG if the polymer chain length is sufficiently small 
