
Alex Deiters
Professor of Chemistry
Research
1) Optochemical control of oligonucleotide function.
We are developing tools that enable light-regulation of DNA and RNA function in cells and animals. Our approach is based on a fundamental concept: the blocking of essential functional groups on biologically active molecules with light-removable protecting groups, so called "caging groups". These caging groups are site-specifically installed, rendering the biological target completely inactive, until the are removed through illumination with UV, visible, or IR light. Since light can be controlled spatially and temporally, the activity of the caged molecule can be precisely controlled in location and timing. In order to achieve light-control of oligonucleotide function, we are developing synthetic approaches to monomeric building blocks for oligonucleotides that contain caged nucleobases. The caging groups block Watson-Crick hydrogen-bonding and completely prevent oligonucleotide duplex formation, until removed through a brief exposure to UV light.
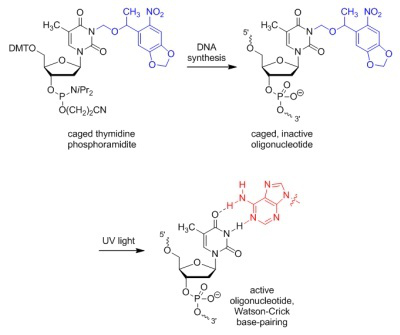
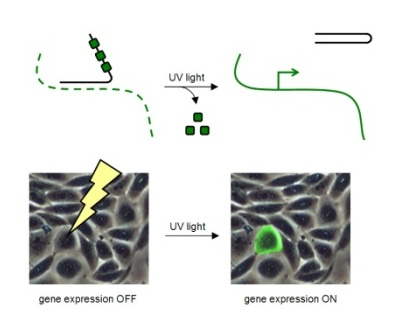
When the caging groups are installed on antisense oligonucleotides, they enable optochemical control of ON to OFF and OFF to ON switching of gene expression with high spatial and temporal resolution in human cells, tissues, and multicellular organisms. When bioconjugated to delivery agents, the caging groups enable targeted delivery of oligonucleotides in addition to spatio-temporal control.
2) Engineering light-switches into DNA and RNA processing enzymes.
In order to site-specifically introduce caging groups onto amino acid residues in proteins, we apply a synthetic biology approach using cells engineered with an expanded genetic code. The protein containing the caged amino acid, typically in an active site location, is expressed inside the cells and is inactive until brief exposure to noninvasive UV light removes the caging group and activates protein function. The cellular process placed under optochemical control can then be studied with high spatial and temporal resolution. For example, an optochemically controlled T7 RNA polymerase was constructed by blocking access to the active site for incoming nucleotide triphosphates through installation of a caging group on a crucial lysine residue. The genetically encoded light-controlled enzyme was completely inactive when expressed in mammalian cells, until a brief UV illumination removed the caging group, thereby allowing access to the active site and activating RNA polymerization. By engineering cells that were expressing the caged polymerase with genes or non-coding RNA sequences under control of a T7 promoter, we were able to achieve optochemical gene activation and light-triggered RNA interference. This enabled spatial control of gene function in mammalian cells, as shown for the localized activation of GFP in mammalian tissue culture. Importantly, only cells that were exposed to UV light display activation of protein function. This approach was also applied to the optical control of a DNA polymerase, Cre recombinase, and a zinc-finger nuclease.

3) Discovery of small molecule modifiers of the microRNA pathway.
We are developing small molecules that target the microRNA pathway. MicroRNAs (miRNAs) are single-stranded noncoding RNAs of approximately 22 nucleotides that represent a new class of gene regulators. It is estimated that >30% of all genes and almost every genetic network are (in part) controlled by miRNAs. Cancer, cardiovascular disease, and several viral infections have all been linked to miRNAs. Thus, developing small molecule modifiers of the miRNA pathway may have significant therapeutic implications. We have discovered the first small molecule inhibitors of the miRNA pathway. For example, we developed a cellular assay for miR-122 function, conducted a pilot screen followed by a high-throughput screen, and have identified two inhibitors, which efficiently silence miR-122 function. We selected miR-122 as a target, since it is the most abundant miRNA in the liver and is involved in hepatitis C virus infection and other liver diseases. In addition, we are developing computational circuits based on DNA hybridization reactions in order to detect patterns of miRNA expression in cancer cells. These computation devices could be applied to the detection of disease and analysis of disease progression.
Publications
Prokup, A.; Deiters, A. “Interfacing Synthetic DNA Logic Operations with Protein Outputs”, Angew. Chem. Int. Ed. 2014, 53, 13192-13195.
Yamazoe,
S.; Liu, Q.; McQuade, L. E.; Deiters, A.; Chen J. K. “Sequential Gene
Silencing Using Wavelength-Selective Caged Morpholinos”, Angew. Chem.
Int. Ed. 2014, 53, 10114-10118.
Hemphill, J.; Govan, J.
M.; Tsang, M.; Deiters, A. "Site-Specific Promoter Caging Enables
Optochemical Gene Activation in Cells and Animals", J. Am. Chem. Soc. 2014, 136, 7152-7158.
Liu, Q.; Deiters, A. "Optochemical Control of Deoxyoligonucleotide Function via a Nucleobase-Caging Approach", Acc. Chem. Res. 2014, 47, 45-55.
Govan, J. M.; Young, D. D.; Lusic, H.; Liu, Q.; Lively, M. O.; Deiters, A. "Optochemical Control of RNA Interference in Mammalian Cells", Nucleic Acid Res. 2013, 41, 10518-10528.
Hemphill, J.; Chou, C.; Chin, J. W., Deiters, A. "Genetically Encoded Light-Activated Transcription for Spatio-Temporal Control of Gene Expression and Gene Silencing in Mammalian Cells", J. Am. Chem. Soc. 2013, 135, 13433-13439.
Govan, J. M.; Uprety, R.; Thomas, M.; Lusic, H.; Lively, M. O.; Deiters, A. "Cellular Delivery and Photochemical Activation of Antisense Agents through a Nucleobase Caging Strategy", ACS Chem. Biol. 2013, 2272-2282.
Hemphill, J.; Deiters, A. "DNA Computation in Mammalian Cells: MicroRNA Logic Operations", J. Am. Chem. Soc. 2013, 135, 10512-10518.
Connelly, C. M.; Uprety, R.; Hemphill, J.; Deiters, A. “Spatiotemporal Control of MicroRNA Function using Light-Activated Antagomirs”, Mol. BioSys. 2012, 8, 2987-2993.
Govan, J. M.; Uprety, R.; Hemphill, J.; Lively, M. O.; Deiters, A. “Regulation of Transcription through Light-Activation and Light-Deactivation of Triplex-Forming Oligonucleotides in Mammalian Cells”,
ACS Chem. Biol. 2012, 7, 1247-1256.
Prokup, A.; Hemphill, J.; Deiters, A. “DNA Computation: A Photochemically Controlled AND Gate”, J. Am. Chem. Soc. 2012, 134, 3810-3815.
Govan, J. M.; McIver, A. L.; Deiters, A. “Stabilization and Photochemical Regulation of Antisense Agents through PEGylation”, Bioconjugate Chem. 2011, 22, 2136-2142.
Govan, J. M.; Lively, M. O.; Deiters, A. “Photochemical Control of DNA Decoy Function Enables Precise Regulation of NF-kB Activity”, J. Am. Chem. Soc. 2011, 133, 13176-13182.
Chou, C.; Deiters, A. "Light-Activated Gene Editing with a Photocaged Zinc-Finger Nuclease", Angew. Chem. Int. Ed. 2011, 50, 6839-6842.